Abstract
Exosomes are membraneous nanovesicles of endocytic origin released by most cell types from diverse organisms; they play a critical role in cell–cell communication. ExoCarta (http://www.exocarta.org) is a manually curated database of exosomal proteins, RNA and lipids. The database catalogs information from both published and unpublished exosomal studies. The mode of exosomal purification and characterization, the biophysical and molecular properties are listed in the database aiding biomedical scientists in assessing the quality of the exosomal preparation and the corresponding data obtained. Currently, ExoCarta (Version 3.1) contains information on 11 261 protein entries, 2375 mRNA entries and 764 miRNA entries that were obtained from 134 exosomal studies. In addition to the data update, as a new feature, lipids identified in exosomes are added to ExoCarta. We believe that this free web-based community resource will aid researchers in identifying molecular signatures (proteins/RNA/lipids) that are specific to certain tissue/cell type derived exosomes and trigger new exosomal studies.
INTRODUCTION
Exosomes are 40–100 nm diameter membrane vesicles of endocytic origin that are released by most cell types under normal and pathological conditions upon fusion of multivesicular bodies (MVBs) with the plasma membrane (PM) (1,2). Since the original observation of exosome secretion from reticulocytes (3,4), the release of these membrane particles in vitro into culture medium is reported in other cells (2,5–7). Exosomes have a cup shaped appearance as visioned by electron microscopy, a buoyant density in sucrose of 1.10–1.21 g/ml; and sediment at 100 000g. Exosomes harbor proteins/RNA/lipids that reflect the functionality of the host cell and posses molecular signatures or footprints resembling the diseased cell from which they were secreted (7). Recent studies have shown that exosomes can be isolated in vivo in bodily fluids such as blood (8), urine (9), breast milk (10), amniotic fluid (11), malignant ascites (12), bronchoalveolar lavage fluid (13) and synovial fluid (14). The accessibility of exosomes in bodily fluids makes them an ideal specimen for biomarker discovery.
While several protocols are employed to isolate and purify exosomes, characterizing them is strongly based on electron microscopy (size and shape) and western blotting for the existence of protein markers that are involved in exosomal biogenesis such as PDCD6IP (Alix), TSG101 and/or exosomal membrane molecules such as CD9, CD63 and CD81. As the number of exosomal studies expands to include more organisms and cell types, new insights are gained on proteins common to all exosomes irrespective of the cell-type origin as well as host–cell-specific proteins (15). A database of proteins, RNA and lipids identified in exosomes will reveal classes of molecules that are more often found in exosomes and those that are specific to certain cell types. Such additional information on host cell specific exosomal membrane molecules will trigger further immunoaffinity-based studies and aid biomedical researchers to isolate disease cell derived exosomes devoid of exosomes secreted by other normal cells. Here we describe, ExoCarta, a manually curated web-based database (http://www.exocarta.org) of the exosomal molecular components such as lipids, proteins, mRNA and miRNA as reported in the scientific literature (16). More importantly, the database contains annotations on the exosomal isolation and purification procedures which will indicate the users on the reliability of the exosome preparation.
ExoCarta DATABASE STRUCTURE AND FUNCTIONALITY
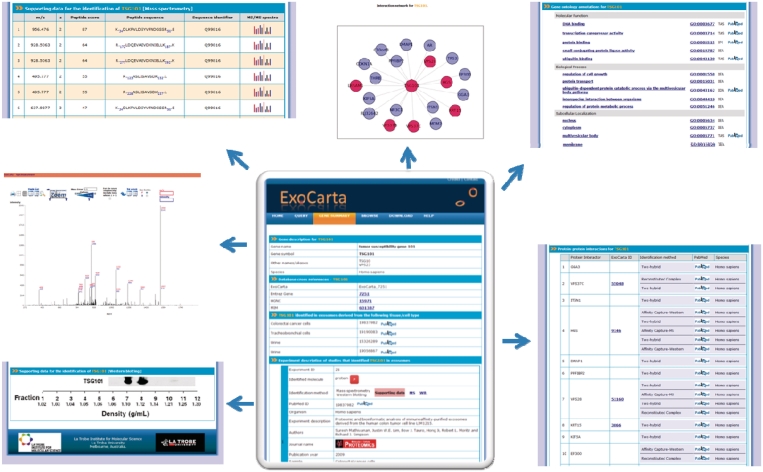
ExoCarta was previously described 2 years ago (16). Briefly, ExoCarta is built with ZOPE, an open source Python-based web application server with dynamic HTML templates, scripts, a search engine and connections for a relational database. Python is used in the three-tier system to connect the web interface with a MySQL database. ExoCarta can be queried using the gene symbol/name or browsed as a set based on the organism, content type (proteins or lipids) and the sample material (plasma or urine). The molecule page in ExoCarta provides the users with molecule centric view on experiment description of the study that identified the molecule, isolation procedures employed, gene ontology-based annotations, protein–protein interactions and a graphical display of the interaction network. Meta annotations regarding the experiment description include the sample from which exosomes were isolated (e.g. plasma, breast cancer cell line), isolation procedures, the identification method (e.g. mass spectrometry), PubMed identifiers, investigators of the study, the journal in which the article was published, the year of the publication. Protein–protein interaction data that was obtained from BioGRID (17) is used in the molecule page to display the protein interaction network graphically as nodes and edges (Figure 1).
Figure 1.
A snapshot of the protein molecule page of TSG101 in ExoCarta is displayed. Users can access molecule centric information from the protein molecule page in ExoCarta. The gene ontology annotations, protein–protein interaction network, graphical display of the network, experimental description pertaining to the studies that identified TSG101, mass spectrometry-based peptide identifications along with the MS/MS spectra and autoradiographs for western blotting are provided wherever possible as supporting information.
ExoCarta DATABASE ADDITIONS AND NEW FEATURES
Data update
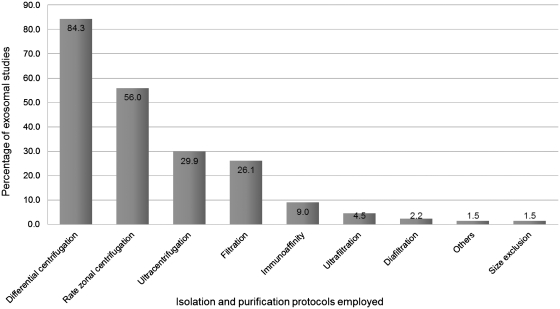
Since the introduction of ExoCarta in April 2009 (16), the exosomal studies added to ExoCarta have doubled. As shown in Table 1, ExoCarta currently comprises 11 261 protein entries resulting from 4049 unique proteins. A total of 4049 mRNA entries and 764 miRNA entries obtained from 134 independent studies are also cataloged in ExoCarta. As homogenous population of exosomes is of paramount importance, the isolation procedures used will indicate the users on the reliability of the exosome preparation and the corresponding data obtained. In this regard, every study in ExoCarta is annotated carefully for the experimental procedures employed to isolate exosomes. Typical isolation procedures such as differential centrifugation, size-based membrane filtration, rate zonal centrifugation, flow cytometry and immunoaffinity pull downs are clearly indicated along with the floatation gradient density of exosomes as reported in the study. The statistical distribution of the isolation protocols used in the studies catalogued in ExoCarta is displayed in Figure 2. Totally, 84% of the studies employed differential centrifugation, 56% used rate zonal centrifugation, 30% used ultracentrifugation and 9% employed immunoaffinity-based methods. It has to be noted that many of the studies used a combination of these procedures. For example, filtration (26%), cannot be used alone to isolate exosomes, was used in conjunction with other isolation procedures. While earlier exosomal studies employed the far simpler approaches of differential centrifugation (15,18), it is encouraging to see that recent exosomal studies are using stringent protocols to isolate exosomes to homogeneity. This is evident by the fact that only 17 of the 134 studies used differential centrifugation alone as the isolation procedure.
Table 1.
Statistics of data cataloged in ExoCarta
| 1 | Number of exosome studies | 134 |
| 2 | Number of protein entries | 11 261 |
| 3 | Number of proteins | 4049 |
| 4 | Number of mRNA entries | 2375 |
| 5 | Number of mRNA molecules | 1639 |
| 6 | Number of lipid molecules | 58 |
| 7 | Number of miRNA molecules | 764 |
| 8 | Number of organisms from which exosomes were isolated | 8 |
| 9 | Number of cell types from which exosomes were isolated | 93 |
| 10 | Number of body fluids from which exosomes were isolated | 12 |
Figure 2.
Distribution of exosomes isolation procedures in the studies catalogued in ExoCarta. A histogram of the distribution of exosome isolation procedures in exosomal studies is displayed. As shown, 84% of the studies employed differential centrifugation, 56% used rate zonal centrifugation and 9% employed immunoaffinity-based methods. It has to be noted that many of the studies used a combination of these procedures. For example, filtration (26%), cannot be used alone to isolate exosomes, was used in conjunction with other isolation procedures.
Addition of lipids
As a new feature of ExoCarta, lipid molecules identified in various exosomal studies are also added in the database. Recent studies on exploring the function/composition of lipids in exosomes have resulted in handful of lipid-based exosomal studies. Currently, ExoCarta catalogs 10 lipid-based exosomal studies. Lipid composition analysis have been performed with exosomes derived from dendritic cells (19), basophilic leukemia cells (20), mast cells (19), reticulocytes (21) and B cells (22). Lipids play a vital role in exosome biogenesis (23) and are characteristic of the cell origin. Interestingly, internal membranes of MVBs are shown to be enriched with lipids such as lyosbisphosphatidic acid (LBPA) (24) which plays an important role in exosome biogenesis, especially ILV formation (23).
Involvement of exosomal community
Manual curation of scientific literature is a cumbersome process. Most of the data present in the scientific literature either as inline text or in the supplementary tables cannot be easily queried upon and are generally lost over time (25). Already existing exosomal datasets in the scientific literature either as inline text or in the supplementary tables are annotated in a communal repository (ExoCarta) with the aid of manual curation. However, in order to keep the database updated, it is very important for the involvement of the exosome scientific community. To facilitate the active involvement of the exosomal scientific community, we have incorporated data contributions with undue credit provided to investigators who generated the data. In this regard, we have instituted a mechanism that allows referees to access exosomal data sets that are submitted by authors before publication in an authorized and anonymous fashion for evaluation purposes. The uploaded data will be highlighted as private and can only be accessed by the authors or their collaborators. Alternatively, the authors can make the data publicly accessible whenever they are inclined to do so. By undertaking such ways of data deposition, the onus on the curators can be reduced several folds. All contributing investigators and the datasets will be listed in ExoCarta credits page (http://www.exocarta.org/credits). Currently, eight investigators have provided their exosomal datasets to the database and we expect more contributions from the exosomal community in the near future.
FUTURE DIRECTIONS
ExoCarta will be updated with more exosomal studies and more fields including protein–protein interactions and post-translational modifications will be incorporated. Microvesicle-based datasets will also be added to ExoCarta in the near future but with clear annotations representing the type/name of the vesicle. ExoCarta will also function as an active news forum in terms of highlighting international scientific meetings and workshops in the context of exosomes or microvesicles. We intend to involve the exosomal scientific community in updating ExoCarta and foresee ExoCarta as a primary resource for exosome research.
DATA USABILITY
ExoCarta is a free to use web-based database. The manually curated data present in ExoCarta (Version 3.1) is available as tab-delimited files and is free for download to all the users.
FUNDING
National Health and Medical Research Council, Australia (Program grant 487922 to R.J.S.); NH&MRC fellowship (1016599 to S.M.). Funding for open access charge: National Health and Medical Research Council, Australia (Program grant 487922 to R.J.S.).
Conflict of interest statement. None declared.
REFERENCES
- 1.Pan BT, Johnstone RM. Fate of the transferrin receptor during maturation of sheep reticulocytes in vitro: selective externalization of the receptor. Cell. 1983;33:967–978. doi: 10.1016/0092-8674(83)90040-5. [DOI] [PubMed] [Google Scholar]
- 2.Simpson RJ, Lim JW, Moritz RL, Mathivanan S. Exosomes: proteomic insights and diagnostic potential. Expert Rev. Proteomics. 2009;6:267–283. doi: 10.1586/epr.09.17. [DOI] [PubMed] [Google Scholar]
- 3.Johnstone RM. Exosomes biological significance: a concise review. Blood Cells Mol. Dis. 2006;36:315–321. doi: 10.1016/j.bcmd.2005.12.001. [DOI] [PubMed] [Google Scholar]
- 4.Vidal M, Mangeat P, Hoekstra D. Aggregation reroutes molecules from a recycling to a vesicle-mediated secretion pathway during reticulocyte maturation. J. Cell Sci. 1997;110(Pt 16):1867–1877. doi: 10.1242/jcs.110.16.1867. [DOI] [PubMed] [Google Scholar]
- 5.Raposo G, Nijman HW, Stoorvogel W, Liejendekker R, Harding CV, Melief CJ, Geuze HJ. B lymphocytes secrete antigen-presenting vesicles. J. Exp. Med. 1996;183:1161–1172. doi: 10.1084/jem.183.3.1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zitvogel L, Regnault A, Lozier A, Wolfers J, Flament C, Tenza D, Ricciardi-Castagnoli P, Raposo G, Amigorena S. Eradication of established murine tumors using a novel cell-free vaccine: dendritic cell-derived exosomes. Nat. Med. 1998;4:594–600. doi: 10.1038/nm0598-594. [DOI] [PubMed] [Google Scholar]
- 7.Mathivanan S, Lim JW, Tauro BJ, Ji H, Moritz RL, Simpson RJ. Proteomics analysis of A33 immunoaffinity-purified exosomes released from the human colon tumor cell line LIM1215 reveals a tissue-specific protein signature. Mol. Cell. Proteomics. 2010;9:197–208. doi: 10.1074/mcp.M900152-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Caby MP, Lankar D, Vincendeau-Scherrer C, Raposo G, Bonnerot C. Exosomal-like vesicles are present in human blood plasma. Int. Immunol. 2005;17:879–887. doi: 10.1093/intimm/dxh267. [DOI] [PubMed] [Google Scholar]
- 9.Pisitkun T, Shen RF, Knepper MA. Identification and proteomic profiling of exosomes in human urine. Proc. Natl Acad. Sci. USA. 2004;101:13368–13373. doi: 10.1073/pnas.0403453101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Admyre C, Johansson SM, Qazi KR, Filen JJ, Lahesmaa R, Norman M, Neve EP, Scheynius A, Gabrielsson S. Exosomes with immune modulatory features are present in human breast milk. J. Immunol. 2007;179:1969–1978. doi: 10.4049/jimmunol.179.3.1969. [DOI] [PubMed] [Google Scholar]
- 11.Taylor DD, Akyol S, Gercel-Taylor C. Pregnancy-associated exosomes and their modulation of T cell signaling. J. Immunol. 2006;176:1534–1542. doi: 10.4049/jimmunol.176.3.1534. [DOI] [PubMed] [Google Scholar]
- 12.Bard MP, Hegmans JP, Hemmes A, Luider TM, Willemsen R, Severijnen LA, van Meerbeeck JP, Burgers SA, Hoogsteden HC, Lambrecht BN. Proteomic analysis of exosomes isolated from human malignant pleural effusions. Am. J. Respir. Cell Mol. Biol. 2004;31:114–121. doi: 10.1165/rcmb.2003-0238OC. [DOI] [PubMed] [Google Scholar]
- 13.Admyre C, Grunewald J, Thyberg J, Gripenback S, Tornling G, Eklund A, Scheynius A, Gabrielsson S. Exosomes with major histocompatibility complex class II and co-stimulatory molecules are present in human BAL fluid. Eur. Respir. J. 2003;22:578–583. doi: 10.1183/09031936.03.00041703. [DOI] [PubMed] [Google Scholar]
- 14.Skriner K, Adolph K, Jungblut PR, Burmester GR. Association of citrullinated proteins with synovial exosomes. Arthritis Rheum. 2006;54:3809–3814. doi: 10.1002/art.22276. [DOI] [PubMed] [Google Scholar]
- 15.Mathivanan S, Ji H, Simpson RJ. Exosomes: extracellular organelles important in intercellular communication. J. Proteomics. 2010;73:1907–1920. doi: 10.1016/j.jprot.2010.06.006. [DOI] [PubMed] [Google Scholar]
- 16.Mathivanan S, Simpson RJ. ExoCarta: a compendium of exosomal proteins and RNA. Proteomics. 2009;21:4997–5000. doi: 10.1002/pmic.200900351. [DOI] [PubMed] [Google Scholar]
- 17.Breitkreutz BJ, Stark C, Reguly T, Boucher L, Breitkreutz A, Livstone M, Oughtred R, Lackner DH, Bahler J, Wood V, et al. The BioGRID Interaction Database: 2008 update. Nucleic Acids Res. 2008;36:D637–D640. doi: 10.1093/nar/gkm1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Welton JL, Khanna S, Giles PJ, Brennan P, Brewis IA, Staffurth J, Mason MD, Clayton A. Proteomics analysis of bladder cancer exosomes. Mol. Cell. Proteomics. 2010;9:1324–1338. doi: 10.1074/mcp.M000063-MCP201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Laulagnier K, Motta C, Hamdi S, Roy S, Fauvelle F, Pageaux JF, Kobayashi T, Salles JP, Perret B, Bonnerot C, et al. Mast cell- and dendritic cell-derived exosomes display a specific lipid composition and an unusual membrane organization. Biochem. J. 2004;380:161–171. doi: 10.1042/BJ20031594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Subra C, Grand D, Laulagnier K, Stella A, Lambeau G, Paillasse M, De Medina P, Monsarrat B, Perret B, Silvente-Poirot S, et al. Exosomes account for vesicle-mediated transcellular transport of activatable phospholipases and prostaglandins. J. Lipid Res. 2010;51:2105–2120. doi: 10.1194/jlr.M003657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vidal M, Sainte-Marie J, Philippot JR, Bienvenue A. Asymmetric distribution of phospholipids in the membrane of vesicles released during in vitro maturation of guinea pig reticulocytes: evidence precluding a role for “aminophospholipid translocase”. J. Cell. Physiol. 1989;140:455–462. doi: 10.1002/jcp.1041400308. [DOI] [PubMed] [Google Scholar]
- 22.Wubbolts R, Leckie RS, Veenhuizen PT, Schwarzmann G, Mobius W, Hoernschemeyer J, Slot JW, Geuze HJ, Stoorvogel W. Proteomic and biochemical analyses of human B cell-derived exosomes. Potential implications for their function and multivesicular body formation. J. Biol. Chem. 2003;278:10963–10972. doi: 10.1074/jbc.M207550200. [DOI] [PubMed] [Google Scholar]
- 23.Chu Z, Witte DP, Qi X. Saposin C-LBPA interaction in late-endosomes/lysosomes. Exp. Cell. Res. 2005;303:300–307. doi: 10.1016/j.yexcr.2004.09.029. [DOI] [PubMed] [Google Scholar]
- 24.Kobayashi T, Gu F, Gruenberg J. Lipids, lipid domains and lipid-protein interactions in endocytic membrane traffic. Semin. Cell Dev. Biol. 1998;9:517–526. doi: 10.1006/scdb.1998.0257. [DOI] [PubMed] [Google Scholar]
- 25.Mathivanan S, Ahmed M, Ahn NG, Alexandre H, Amanchy R, Andrews PC, Bader JS, Balgley BM, Bantscheff M, Bennett KL, et al. Human Proteinpedia enables sharing of human protein data. Nat. Biotechnol. 2008;26:164–167. doi: 10.1038/nbt0208-164. [DOI] [PubMed] [Google Scholar]