Abstract
SMART (Simple Modular Architecture Research Tool) is an online resource (http://smart.embl.de/) for the identification and annotation of protein domains and the analysis of protein domain architectures. SMART version 7 contains manually curated models for 1009 protein domains, 200 more than in the previous version. The current release introduces several novel features and a streamlined user interface resulting in a faster and more comfortable workflow. The underlying protein databases were greatly expanded, resulting in a 2-fold increase in number of annotated domains and features. The database of completely sequenced genomes now includes 1133 species, compared to 630 in the previous release. Domain architecture analysis results can now be exported and visualized through the iTOL phylogenetic tree viewer. ‘metaSMART’ was introduced as a novel subresource dedicated to the exploration and analysis of domain architectures in various metagenomics data sets. An advanced full text search engine was implemented, covering the complete annotations for SMART and Pfam domains, as well as the complete set of protein descriptions, allowing users to quickly find relevant information.
INTRODUCTION
The SMART database (http://smart.embl.de) is now in its 13th year (1), and provides high quality, manually curated Hidden–Markov models and alignments of protein domain families. Accessible though a web interface or via various programmatic methods, SMART remains a popular tool for domain annotation and exploration of protein domain architectures, with an average of 200 000 user submitted proteins analyzed monthly.
IMPROVED DOMAIN COVERAGE
Even though the rate of novel domain discovery is constantly declining (2), SMART gradually expands its domain coverage in each release. The current version 7 introduces more than 200 new domains, bringing the total to 1009 distinct modules that can be searched. Even though many of these domains were already annotated in other databases, like Pfam (3), SMART's domain annotation pipeline relies heavily on manual intervention, making the re-annotation process worthwhile.
UPDATED PROTEIN DATABASES
The number of annotated protein sequences is constantly growing, at the same time increasing the redundancy in the databases. Since protein redundancy significantly skews the number of domains reported in both domain architecture analyses and when comparing domain counts in complete genomes, past versions of SMART (4) introduced several features to minimize these problems. The standard protein database used by SMART combines the complete Uniprot protein database (5) with predicted proteins from all stable Ensembl (6) genomes. Since these are inherently highly redundant, SMART implements a per-species clustering method (7) to minimize the redundancy in the final database. Yet, the updated version currently contains more than 11 million proteins from around 150 thousand species, subspecies and varietas. Additionally, SMART offers a ‘genomic’ analysis mode that contains only proteins from completely sequenced genomes. Synchonized with STRING version 9 (8), this database has been significantly expanded, and contains 1133 complete genomes (121 Eukaryota, 943 Bacteria and 69 Archaea).
NOVEL ARCHITECTURE ANALYSIS DATA EXPORT AND VISUALIZATION FEATURES
Domain architecture analysis functions in SMART allow users to simply access proteins containing combinations of particular domains. These can be also generated using combinations of GO terms (9) associated to protein domains, and restricted to various taxonomic classes. Previous versions of SMART allowed users to download these selected proteins as FASTA formatted files or to display them through schematic representations (SMART ‘bubblograms’). SMART 7 offers a new data export functions for domain architecture analysis, which is tightly coupled with iTOL [interactive Tree Of Life (10,11)], our phylogenetic tree visualization tool.
Data are exported into two separate files, which can be directly used by iTOL: a Newick formatted phylogenetic tree and a protein domain data set file used to visualize proteins on the tree. The procedure is as follows:
an initial list of proteins is obtained through an architecture analysis query;
proteins are grouped according to their species of origin;
these species are used to ‘prune’ the complete NCBI taxonomy database (12) by walking the taxonomy tree up to the root and exporting the resulting structure into a Newick formatted phylogenetic tree; and
each protein's domain organization is converted into a plain text format understood by iTOL.
Resulting plain text files can be downloaded, or directly visualized in iTOL by a simple button click (Figure 1).
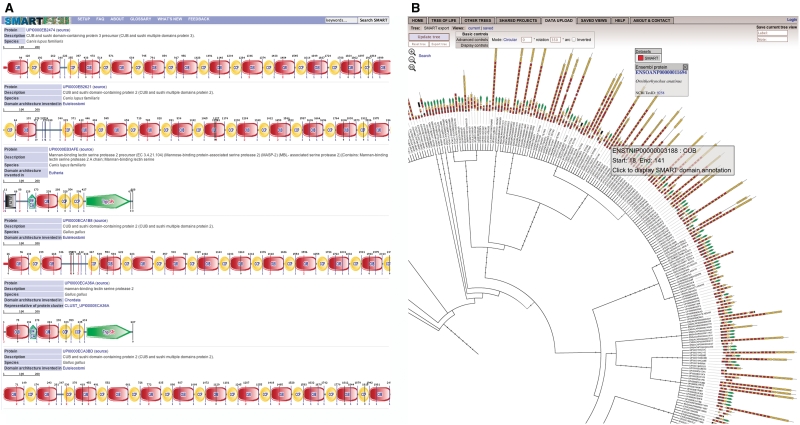
Figure 1.
Displaying SMART protein domain architectures in iTOL. New data export features allow users to simply display domain architecture query results on a NCBI taxonomy based phylogenetic tree. Phylogenetic trees are generated on-the-fly by pruning the NCBI taxonomy database (12) and visualized in interactive Tree Of Life (10). (a) SMART was queried for all proteins containing both CUB and CCP domains. (b) Query results visualized on a phylogenetic tree in iTOL.
EXPANDED PROTEIN INTERACTION DATA
Similar to previous SMART updates, we synchronized our underlying protein interaction data with the latest version of the STRING database (8). Since the number of species in our protein database based on completely sequenced genomes increased almost 2-fold in this release, the information on putative protein interaction partners has also been significantly expanded, and is now available for more than 3.5 million proteins. Interaction network data display has been updated, and uses a streamlined graphical representation, which brings several extra layers of information while being easier to interpret.
metaSMART: BASIC INTEGRATION OF ENVIRONMENTAL SEQUENCING DATA
Metagenomics projects (that is environmental shotgun sequencing) are constantly increasing the amount of novel, uncharacterized DNA and (fragments of) protein sequences. Functional characterization and annotation of such data remains a daunting task, and various pipelines, such as SmashCommunity (13), are being developed to help scientists in this process.
As an initial step toward meaningful integration of these data into SMART, we created ‘metaSMART’. Its primary goal is the exploration and analysis of protein domain architectures in various publicly available metagenomics data sets.
Users can compare different domain frequencies, co-occurrences and complex architectures in different environments to illustrate the role of domain variability depending on the habitat. Furthermore, metaSMART allows the exploration of completely novel domain architectures, unique in databases so far; analyses of various non-described domain compositions could broaden the knowledge about new protein functions related to their domain interdependency (Figure 2). Four metagenomics data sets are the starting point of metaSMART: Sargasso sea (14), acid mine drainage biofilm (15), Minnesota farm soil (16) and ‘Whale fall’ carcasses (16). We are currently integrating several additional metagenomes [for example, the human gut (17)], which will significantly expand the amount of available information in metaSMART and provide novel biological insights in the context of metagenomics.
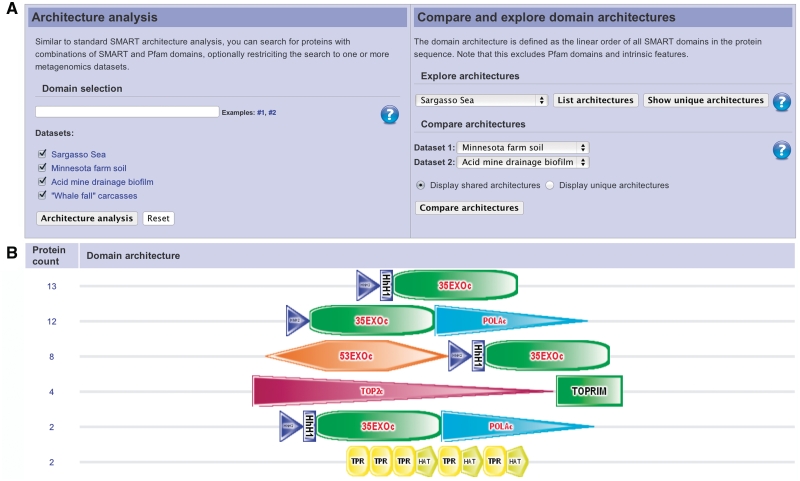
Figure 2.
metaSMART, a novel sub resource dedicated to the exploration of domain architectures in metagenomics data sets. (a) metaSMART user interface provides simple access to all available functions. (b) A subset of protein domain architectures present in the Sargasso Sea data set (14). These are not present in other metagenomics data sets or the standard SMART database, and could be pointing to novel functional associations of various domains.
DATABASE AND WEB SERVER OPTIMIZATIONS
The backend of SMART is a PostgreSQL-based relational database management system, which stores the annotation of all SMART domains and the pre-calculated protein analyses for the entire Uniprot (18), Ensembl (19) and STRING (8) sequence databases. These include SMART and Pfam domains, as well as several protein intrinsic features, like signal peptides, transmembrane and coiled-coil regions. With close to 50 million annotated features in the current database, we have to constantly find new ways of keeping the response times of the server acceptable. Therefore, the database was restructured and several parts of the database access code have been optimized. Additionally, the hardware cluster that powers the sequence annotation searches and database queries has been refreshed and expanded with additional CPUs.
USER INTERFACE IMPROVEMENTS
Version 7 brings various updates to SMART's web interface. Many parts of the interface have been simplified and compacted, resulting in easier navigation and simpler identification of relevant content. To make SMART more accessible to new users, we added help popup windows to various parts of the interface, making different functions easier to understand.
A new full text search engine has been implemented, based on KinoSearch libraries (http://incubator.apache.org/lucy). It indexes the complete annotation pages for all SMART and Pfam domains, as well as Uniprot, Ensembl and STRING protein descriptions, allowing users to quickly identify domains or proteins of interest.
Programmatic access to SMART has been extended with easy to parse text-only output mode, allowing simple batch access to the SMART search engine. Ready to use example scripts that use the batch access interface are also provided.
FUNDING
EMBL (internal budget) and the European Union under the program ‘FP7 capacities: Scientific Data Repositories’ (grant 213037) (IMproving Protein Annotation and Co-ordination using Technology – IMPACT). Funding for open access charge: EMBL (internal budget).
Conflict of interest statement. None declared.
REFERENCES
- 1.Schultz J, Milpetz F, Bork P, Ponting CP. SMART, a simple modular architecture research tool: identification of signaling domains. Proc. Natl Acad. Sci. USA. 1998;95:5857–5864. doi: 10.1073/pnas.95.11.5857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Heger A, Holm L. Exhaustive enumeration of protein domain families. J. Mol. Biol. 2003;328:749–767. doi: 10.1016/s0022-2836(03)00269-9. [DOI] [PubMed] [Google Scholar]
- 3.Finn RD, Mistry J, Tate J, Coggill P, Heger A, Pollington JE, Gavin OL, Gunasekaran P, Ceric G, Forslund K, et al. The Pfam protein families database. Nucleic Acids Res. 2010;38:D211–D222. doi: 10.1093/nar/gkp985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Letunic I, Copley RR, Pils B, Pinkert S, Schultz J, Bork P. SMART 5: domains in the context of genomes and networks. Nucleic Acids Res. 2006;34:D257–D260. doi: 10.1093/nar/gkj079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Consortium TU. The Universal Protein Resource (UniProt) in 2010. Nucleic Acids Res. 2010;38:D142–D148. doi: 10.1093/nar/gkp846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Flicek P, Amode MR, Barrell D, Beal K, Brent S, Chen Y, Clapham P, Coates G, Fairley S, Fitzgerald S, et al. Ensembl 2011. Nucleic Acids Res. 2011;39:D800–D806. doi: 10.1093/nar/gkq1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Letunic I, Doerks T, Bork P. SMART 6: recent updates and new developments. Nucleic Acids Res. 2009;37:D229–D232. doi: 10.1093/nar/gkn808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Szklarczyk D, Franceschini A, Kuhn M, Simonovic M, Roth A, Minguez P, Doerks T, Stark M, Muller J, Bork P, et al. The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res. 2011;39:D561–D568. doi: 10.1093/nar/gkq973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blake JA, Harris MA. The Gene Ontology (GO) project: structured vocabularies for molecular biology and their application to genome and expression analysis. Curr. Protoc. Bioinformatics. 2008 doi: 10.1002/0471250953.bi0702s23. Chapter 7, Unit 7 2. [DOI] [PubMed] [Google Scholar]
- 10.Letunic I, Bork P. Interactive Tree Of Life v2: online annotation and display of phylogenetic trees made easy. Nucleic Acids Res. 2011;39:W475–W478. doi: 10.1093/nar/gkr201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Letunic I, Bork P. Interactive Tree Of Life (iTOL): an online tool for phylogenetic tree display and annotation. Bioinformatics. 2007;23:127–128. doi: 10.1093/bioinformatics/btl529. [DOI] [PubMed] [Google Scholar]
- 12.Sayers EW, Barrett T, Benson DA, Bolton E, Bryant SH, Canese K, Chetvernin V, Church DM, DiCuccio M, Federhen S, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2011;39:D38–D51. doi: 10.1093/nar/gkq1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Arumugam M, Harrington ED, Foerstner KU, Raes J, Bork P. SmashCommunity: a metagenomic annotation and analysis tool. Bioinformatics. 2010;26:2977–2978. doi: 10.1093/bioinformatics/btq536. [DOI] [PubMed] [Google Scholar]
- 14.Venter JC, Remington K, Heidelberg JF, Halpern AL, Rusch D, Eisen JA, Wu D, Paulsen I, Nelson KE, Nelson W, et al. Environmental genome shotgun sequencing of the Sargasso Sea. Science. 2004;304:66–74. doi: 10.1126/science.1093857. [DOI] [PubMed] [Google Scholar]
- 15.Tyson GW, Chapman J, Hugenholtz P, Allen EE, Ram RJ, Richardson PM, Solovyev VV, Rubin EM, Rokhsar DS, Banfield JF. Community structure and metabolism through reconstruction of microbial genomes from the environment. Nature. 2004;428:37–43. doi: 10.1038/nature02340. [DOI] [PubMed] [Google Scholar]
- 16.Tringe SG, von Mering C, Kobayashi A, Salamov AA, Chen K, Chang HW, Podar M, Short JM, Mathur EJ, Detter JC, et al. Comparative metagenomics of microbial communities. Science. 2005;308:554–557. doi: 10.1126/science.1107851. [DOI] [PubMed] [Google Scholar]
- 17.Arumugam M, Raes J, Pelletier E, Le Paslier D, Yamada T, Mende DR, Fernandes GR, Tap J, Bruls T, Batto JM, et al. Enterotypes of the human gut microbiome. Nature. 2011;473:174–180. doi: 10.1038/nature09944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Consortium TU. The universal protein resource (UniProt) Nucleic Acids Res. 2008;36:D190–D195. doi: 10.1093/nar/gkm895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Flicek P, Aken BL, Beal K, Ballester B, Caccamo M, Chen Y, Clarke L, Coates G, Cunningham F, Cutts T, et al. Ensembl 2008. Nucleic Acids Res. 2008;36:D707–D714. doi: 10.1093/nar/gkm988. [DOI] [PMC free article] [PubMed] [Google Scholar]