Abstract
With a worldwide prevalence of ∼5%, attention deficit hyperactivity disorder (ADHD) has become one of the most common psychiatric disorders. The polygenetic nature of ADHD indicates that multiple genes jointly contribute to the development of this complex disease. Studies aiming to explore genetic susceptibility of ADHD have been increasing in recent years. There is a growing need to integrate the genetic data from various genetic studies to provide a comprehensive data set and uniform access for convenience of in-depth data mining. So far, there has been no such effort for ADHD. To address the genetic complexity of ADHD, we developed the ADHDgene database by integrating ADHD-related genetic factors by profound literature reading. Based on the data from the literature, extended functional analysis, including linkage disequilibrium analysis, pathway-based analysis and gene mapping were performed to provide new insights into genetic causes of ADHD. Moreover, powerful search tools and a graphical browser were developed to facilitate the navigation of the data and data connections. As the first genetic database for ADHD, ADHDgene aims to provide researchers with a central genetic resource and analysis platform for ADHD and is freely available at http://adhd.psych.ac.cn/.
INTRODUCTION
Attention deficit hyperactivity disorder (ADHD) is one of the most common psychiatric disorders with a worldwide prevalence of ∼5% (1). The prevalence is even higher among school-age children ranging from 8% to 12% (2,3). ADHD is predominantly childhood-onset and can persist into adolescence and adulthood to inflict long-term harm. It is characterized by a continuous and combined pattern of inattentive, hyperactive and impulsive behavior, and is often comorbid with other psychiatric disorders (4). Patients with ADHD have impaired academic, executive and social functions, which also lead to a serious financial burden to families and society. Although the etiology of ADHD is still incompletely understood, results from family, twin and adoption studies, as well as molecular genetic studies consistently indicate the strong genetic influence on ADHD with estimated heritability ranging from 75% to 91% (5). To date, the polygenetic nature of ADHD is widely acknowledged, which indicates multiple genes of moderate effect are involved in the genetic basis of ADHD (6). Meanwhile, alternative modes of inheritance in ADHD remain a topic for further clarification. For example, evidences from the segregation analysis demonstrated that a few major genes convey susceptibility to ADHD and they exhibit Mendelian segregation (7). Thus, to study the genetic basis of ADHD is of fundamental importance in uncovering disease mechanism and in developing effective methods for ADHD diagnosis, treatment and prevention.
During the past years, numerous studies aiming to explore genetic susceptibility of ADHD have been published. Large numbers of association studies (8), linkage studies (9) and meta-analyses (10) have been conducted, and numbers of susceptibility variants, genes and chromosomal regions have been reported to be associated with ADHD. For example, both genome-wide linkage and fine mapping studies support the linkage between ADHD and chromosome bands 16p13 (11,12). Genes of the dopaminergic and serotonergic system, including DRD4, SLC6A3 (DAT1) and DBH, were widely studied and implicated to be associated with the susceptibility of ADHD. Several genome-wide association studies (GWAS) have been conducted (13), and studies of genomic DNA copy number variants (CNVs) in ADHD have also emerged in recent years and identified some rare or large deletions/duplications in ADHD patients (14–16). However, these results are scattered in numerous publications and sometimes are equivocal or inconsistent. For example, a candidate–gene association study using a sample of 674 families indicated that both genes, DRD4 and SLC6A3, are associated with ADHD respectively (17), while results from a case–control study did not find any association between these two genes and ADHD (18). As these inconsistent results are common in genetic studies, it would be difficult for researchers to acquire a global understanding of these positive and negative findings, and then design the right kind of study to move forward. The difficulty will be significantly increased with the fast growth of genetic data. Thus, to address the genetic complexity of ADHD and the heterogeneity of studies, a comprehensive and well-organized collection of genetic data from multiple published studies is urgently needed. Moreover, an association study, as the currently most favored and widely used approach for genetic study, usually identifies trait-associated genetic markers rather than causal variants affecting the trait. Regular data analysis of an association study examines SNPs/genes independently and ignores the combined SNP/gene effects. In order to take the best advantage of the current genetic data, in-depth data mining is urgently needed to provide new candidates and new insights into the mechanism of ADHD.
Here, we present ADHDgene, a genetic database for ADHD, to fulfill the growing needs of data integration and data analysis. Currently, there are several genetic databases focusing on psychiatric disorders, like the database of genetic studies of bipolar disorder (19), SzGene (20) and SZGR (21) for schizophrenia, and AutDB (22) and AGD (23) for autism. However, there has been no such effort for ADHD. As the first genetic database for ADHD, ADHDgene aims to provide the research community with a comprehensive set of genetic data from both profound literature screening and extended functional analysis. ADHDgene also provides online tools to search and browse different data types and data connections. In general, ADHDgene is designed to be a central genetic database for ADHD to facilitate the study of function and mechanism to help unveil the genetic basis of ADHD.
DATA CONTENT, DATA INTEGRATION AND ANALYSIS
Data content
The key data types in ADHDgene include variants (SNP, CNV, etc.), genes and chromosomal regions. There are mainly two sets of data hosted by ADHDgene. One is the core data, which was derived from full-text literature reading with manual curation of ADHD genetic studies from the PubMed database at NCBI (http://www.ncbi.nih.gov/pubmed). The other is the extended data derived from extended functional analysis based on the core data, including linkage disequilibrium (LD) analysis of the literature-origin SNPs, pathway-based analysis (PBA) for the GWAS of ADHD and gene mapping. A full annotation was made for both data sets, including SNP functional annotation (such as non-synonymous coding SNPs, or SNPs leading to gain or lost of stop codon) and GO and KEGG pathway annotation for genes. Public databases utilized for the mapping and annotation include dbSNP (24), Ensembl (25), HGNC (26), UCSC (27), GO (28) and KEGG (29). The data statistics of ADHDgene dated 1 August 2011 is shown in Table 1. All data can be freely downloaded from our website.
Table 1.
ADHDgene data content and statistics as of 1 August 2011
| Data seta | Data type | Data statisticsb | |
|---|---|---|---|
| Core data | Variant | SNP | 941 (291) |
| CNV | 296 (NA) | ||
| Othersc | 160 (41) | ||
| Gene | 213 (83) | ||
| Region | 127 (29) | ||
| Extended data | Variant | LD-proxy | 4120 |
| Gene | Mapped by published SNP | 65 | |
| Mapped by LD-proxy | 140 | ||
| Mapped by CNV | 891 | ||
| Mapped by significant region | 2295 | ||
| In pathway identified by PBA | 260 | ||
| Pathway | Pathway identified by PBA | 8 | |
aThe core data is entirely from the literature, while the extended data is from extended functional analysis
bNumber in parenthesis indicates the number of statistically significant results
cOther variants include VNTR, microsatellite, STR, duplication, SNP without reference SNP ID in dbSNP, etc. CNV, copy number variation; PBA, pathway-based analysis; NA, not applicable.
Core data: integration and curation
The core data of ADHDgene integrates ADHD-related genetic factors from published literature. Besides SNP and CNV, other variants like variable number tandem repeat (VNTR) and microsatellite are also included. To obtain the core data, a comprehensive search of ADHD related genetic publications in PubMed was made by using the following search term: (‘attention deficit hyperactivity disorder’ [Title/Abstract] OR ADHD [Title/Abstract]) AND (gene [Title/Abstract] OR genetic [Title/Abstract] OR genome [Title/Abstract] OR genomic [Title/Abstract]) AND (linkage [Title/Abstract] OR association [Title/Abstract] OR polymorphism [Title/Abstract] OR SNP [Title/Abstract] OR variant [Title/Abstract] OR variation [Title/Abstract] OR mutation [Title/Abstract]). It resulted in ∼780 English publications from the year 2000 through 30 June 2011. By manual screening of abstracts of these publications, only publications about association studies, linkage studies and meta-analyses were kept for further reading. Other publications like reviews or articles about pharmacology, sociology, electrophysiology, neurophysiology and behavioral research were excluded. After filtering, there are 288 articles included in ADHDgene. The full text of each eligible publication was read carefully, and detailed information of each study was extracted manually. To ensure a comprehensive review, both positive and negative results were collected.
To illustrate the association between genetic candidates and ADHD, statistical results were further evaluated and were categorized into ‘Significant’, ‘Non-significant’ and ‘Trend’ according to their statistical evidence in the original publications. For linkage study, the significance levels were assigned based on the criteria proposed by Lander and Kruglyak (30). For candidate–gene association studies, a significance level of P < 0.05 was used unless the authors suggested some other value. The result of statistical analysis is defined as ‘Significant’ if the corresponding P-value is <0.05. For GWAS, P < 1 × 10−8 indicates a ‘Significant’ result, P > 1 × 10−5 indicates a ‘Non-significant’ result, and a value between these boundaries represents a ‘Trend’ result. Mutations identified in ADHD patients were all classified as ‘Trend’ unless statistical significance was proposed. Meanwhile, to help understand the results from statistical analysis, for each study, the study design, sample population, analytical method, as well as brief comments from authors, were presented to provide researchers a clear picture for each genetic factor.
Extended data: extended functional analysis
The polygenetic nature of ADHD indicates that multiple genes jointly contribute to the development of ADHD with each having a modest effect on the overall risk (31). Although current research has reported many candidate SNPs and genes for ADHD, few are conclusive due to the low replication rate, or lack of functional evidence on mechanism through which the susceptibility genes jointly affect the trait. With the purpose to identify more candidate SNPs and genes, as well as to allow a deeper interpretation of current data, we performed extended functional analysis based on the core data, which includes LD analysis, pathway-based analysis and gene mapping. These analyses enriched the content of the database to provide new clues for understanding the pathological mechanism of ADHD.
Linkage disequilibrium (LD) analysis
Association studies have identified many SNPs as candidate genetic markers for ADHD, but most of them lack function, e.g. some intronic SNPs, flanking SNPs or synonymous coding SNPs. Linkage disequilibrium (LD) analysis searches the LD-proxies of the literature-origin SNPs with the purpose to capture more candidate SNPs, especially those candidate causal SNPs based on functional annotation. These extended LD-proxies will provide new candidate SNPs for future study designs.
The LD data used in this analysis were downloaded from the HapMap website, which was compiled from merged genotype data from phases I+II+III (HapMap rel #27, NCBI B36) for markers up to 200 kb apart (32). SNPs in LD (r2 > 0.8) with published SNPs were defined as LD-proxies. The population used in the LD analysis is consistent with original studies. For example, the LD-proxies of SNP rs2873804, which was identified in the Chinese population, were extracted from HapMap LD data in the CHB, CHD and JPT populations (32).
Pathway-based analysis (PBA) for genome-wide association study (GWAS)
So far, five genome-wide association studies (GWASs) have been conducted for ADHD but few significant SNPs were found (33–37). As the original GWAS analysis focuses on single SNP/gene, combined SNP/gene effects are hardly explored, which leads to difficulties in understanding the biological function and mechanism of complex diseases (38). To overcome this limitation, we implemented a pathway-based analysis (PBA) algorithm, named i-GSEA (improved gene set enrichment analysis) developed in our recent studies (39,40), on the full list of GWAS SNP P-values to detect pathways/gene sets associated with ADHD. i-GSEA utilizes a re-scaled enrichment score (ES) to emphasize pathways with well evaluated high proportion of significant genes to identify pathways/gene sets with combined effects of modest genes involved in complex disease. False discovery rate (FDR) is calculated for each pathway/gene set and pathways/gene sets with FDR < 0.05 are regarded as statistical significance.
The ADHD GWAS data sets (full list of SNP P-values) were requested by emailing the corresponding authors of related publications or through dbGaP (41). Finally, two GWAS data sets were authorized. One is from a family-based GWAS using SNP array (35), and the other is from a case–control GWAS with pooled DNA (34). i-GSEA was performed on the above two data sets by using default parameters (39) with annotated GO terms from MSigDB 3.0 (42) as reference database. In total, the PBA analyses resulted in 8 pathways/gene sets significantly associated with ADHD (FDR < 0.05). Among them, the pathway annotated by GO term GO:0005262 represents a set of genes related to calcium channel activity, which has been reported to infer the etiology of other psychiatric disorders (43,44). This result provides new evidence for possible shared biological mechanisms between ADHD and other psychiatric disorders.
Gene mapping
To ensure a comprehensive collection of candidate genes for ADHD, published SNPs, which were not mapped to genes in the original publications, and LD-proxies from LD analysis were mapped to genes according to their chromosomal locations. Mapping between genes and the reported CNVs and significant chromosomal regions were performed as well. All genes acquired by the above mappings, as well as genes in pathways identified by PBA, were regarded as the extended candidate genes for ADHD. This part of genes is distinguished as ‘genes from other sources’ in comparison to the ‘literature-origin genes’ from the core data in ADHDgene database.
DATABASE USAGE AND ACCESS
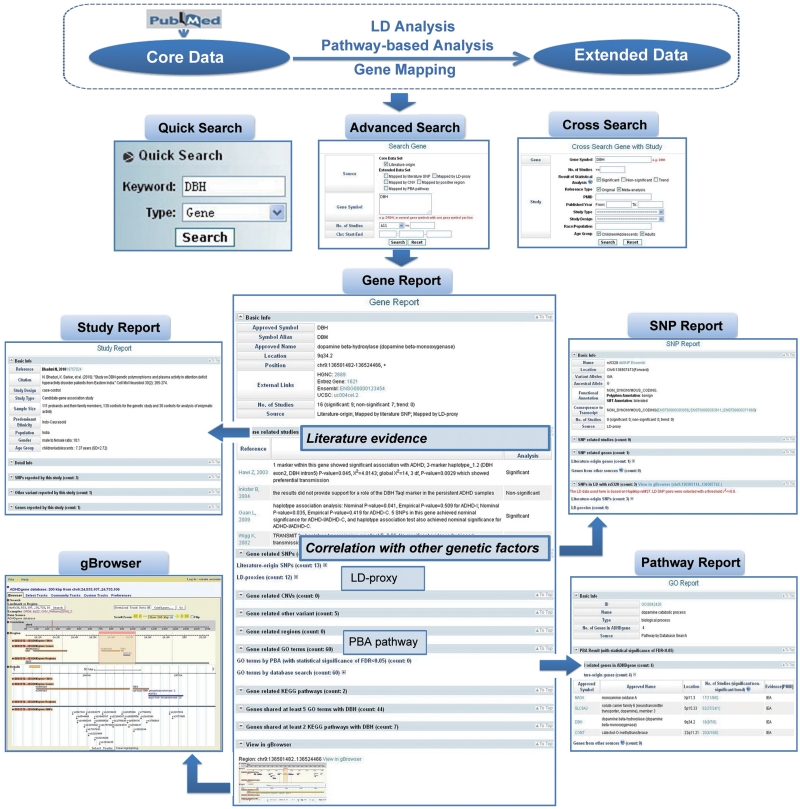
ADHDgene provides researchers with a powerful search engine and a user-friendly interface to access and browse different data types and data connections. To search the database efficiently, besides ‘Quick Search’ by keyword, ADHDgene offers ‘Advanced Search’ for variant, gene, region, pathway and publication to allow users to specify and combine query options. Particularly, to facilitate a thorough investigation on the core data, ADHDgene supports ‘Cross Search’ between different genetic factors (variant, gene and region) and publication, which helps to provide a thorough overview for each genetic factor. For example, users may want to know how many statistically significant associations have been reported for gene dopamine beta-hydroxylase (DBH) in the young generations of Caucasian populations. Through ‘Cross Search’, users may input the gene name ‘DBH’, select ‘Significant’ in the field ‘Result of statistical analysis’, input ‘Caucasian’ in the field ‘Population’ and select ‘Children/Adolescents’ in the field ‘Age group’.
ADHDgene provides a detailed report for each genetic factor. Taking the gene DBH for example, on its ‘Gene Report’, DBH-related 16 publications and key results from each publication are listed. Among them, nine articles reported significant associations with ADHD, and seven reported non-significant associations. To assess the heterogeneity of these studies, users may link to the ‘Publication Report’ to view the details. Other genetic factors related to DBH are all listed, such as SNPs (literature-origin or LD-proxies), CNVs, regions, and genes sharing same pathway with DBH (by database annotation or PBA, which indicate potential candidates for epistasis study). It is worth mentioning that there are 12 LD-proxies identified for DBH. Five of them have been reported to have linkage or association signals with other diseases, like schizophrenia (45,46) and Parkinson’s disease (47). It is likely that these LD-proxies could be possible candidate markers for ADHD.
Besides the search module, GBrowse (48) was incorporated in ADHDgene to facilitate viewing the different types of genetic factors (variant/gene/region) simultaneously in the context of genomic regions. GBrowse is a popular visualization tool for visualizing genetic and genomic data. Entries from the literature and extended functional analysis are marked in different colors in the browser. Users can also link to the detail page of corresponding entries from the browser. In summary, through the above search and browse tools, users can access and view the logical connections between different genetic factors with strong evidence support either from the literature or from functional analysis (Figure 1).
Figure 1.
Screenshot of the search and browse tools for access and analysis of data and data connections in ADHDgene.
SYSTEM DESIGN AND IMPLEMENTATION
All data of ADHDgene are stored and managed in a MySQL relational database. The website is implemented using JSP running on an Apache Tomcat web server. Struts framework and Hibernate were employed to help improve the stability of the web services. AJAX and some jQuery plugins were used for the interface development. The data analysis programs were written by PERL. GBrowse uses MySQL as backend and was built following the configuration files provided by its developer (http://gmod.org/wiki/GBrowse_Configuration_HOWTO). The genomic data in ADHDgene are loaded into GBrowse after being converted into genome feature format (GFF).
DISCUSSION AND FUTURE DEVELOPMENT
As the first genetic database for ADHD, ADHDgene was developed to provide a panoramic view of present genetic studies for this disease. ADHDgene covers a broad range of data types including not only SNPs, genes, chromosomal regions and pathways, but also variants like CNV, VNTR and microsatellites. By integrating data from both publications and extended functional analysis, ADHDgene provides a comprehensive genetic data set for ADHD. In comparison to other disease-centered genetic databases, ADHDgene was developed in a reliable (manual curation of literature) but not limited (extended functional analysis) way to fulfill the increasing research demands in addressing the genetic complexity of ADHD. By providing powerful search and browse tools, the ADHDgene database aims to act as not only an integrated genetic resource for ADHD, but also as a flexible application platform for the genetic study of ADHD.
In the next few years, the number of genetic studies of ADHD is expected to keep increasing especially with the development of new technologies, such as next generation sequencing (49). ADHDgene will be periodically updated to ensure a most up-to-date follow up of the genetic research progress of ADHD. It should be noted that pathways by PBA in ADHDgene provide potential hypotheses for further study rather than conclusive results, since the results from PBA might be affected by the quality of GWAS data (50). As soon as additional high quality GWAS data become available, we will analyze them to update the PBA results in ADHDgene. Meanwhile, as studies of rare variants, epigenetics, large-scale gene expression and gene–environment interplay for ADHD are expected to accelerate, the scope of ADHDgene will be expanded to integrate newly generated data. Evidence from animal models will also be one of the future efforts in updating ADHDgene. We hope our continuous efforts will help to unveil the genetic basis of ADHD and to contribute to global mental health.
FUNDING
Knowledge Innovation Program of the Chinese Academy of Sciences (KSCX2-EW-J-8); National Natural Science Foundation of China (81101545). Funding for open access charge: Knowledge Innovation Program of the Chinese Academy of Sciences (KSCX2-EW-J-8).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank all our colleagues and friends in the Chinese Academy of Sciences and from different Universities both inside and outside China who helped us test the database and provided the valuable suggestions.
REFERENCES
- 1.Polanczyk G, de Lima MS, Horta BL, Biederman J, Rohde LA. The worldwide prevalence of ADHD: a systematic review and metaregression analysis. Am. J. Psychiatry. 2007;164:942–948. doi: 10.1176/ajp.2007.164.6.942. [DOI] [PubMed] [Google Scholar]
- 2.Faraone SV, Sergeant J, Gillberg C, Biederman J. The worldwide prevalence of ADHD: is it an American condition? World Psychiatry. 2003;2:104–113. [PMC free article] [PubMed] [Google Scholar]
- 3.Biederman J, Faraone SV. Attention-deficit hyperactivity disorder. Lancet. 2005;366:237–248. doi: 10.1016/S0140-6736(05)66915-2. [DOI] [PubMed] [Google Scholar]
- 4.Biederman J, Newcorn J, Sprich S. Comorbidity of attention deficit hyperactivity disorder with conduct, depressive, anxiety, and other disorders. Am. J. Psychiatry. 1991;148:564–577. doi: 10.1176/ajp.148.5.564. [DOI] [PubMed] [Google Scholar]
- 5.Faraone SV, Perlis RH, Doyle AE, Smoller JW, Goralnick JJ, Holmgren MA, Sklar P. Molecular genetics of attention-deficit/hyperactivity disorder. Biol. Psychiatry. 2005;57:1313–1323. doi: 10.1016/j.biopsych.2004.11.024. [DOI] [PubMed] [Google Scholar]
- 6.Tripp G, Wickens JR. Neurobiology of ADHD. Neuropharmacology. 2009;57:579–589. doi: 10.1016/j.neuropharm.2009.07.026. [DOI] [PubMed] [Google Scholar]
- 7.Acosta MT, Arcos-Burgos M, Muenke M. Attention deficit/hyperactivity disorder (ADHD): complex phenotype, simple genotype? Genet. Med. 2004;6:1–15. doi: 10.1097/01.gim.0000110413.07490.0b. [DOI] [PubMed] [Google Scholar]
- 8.Gizer IR, Ficks C, Waldman ID. Candidate gene studies of ADHD: a meta-analytic review. Hum. Genet. 2009;126:51–90. doi: 10.1007/s00439-009-0694-x. [DOI] [PubMed] [Google Scholar]
- 9.Zhou K, Dempfle A, Arcos-Burgos M, Bakker SC, Banaschewski T, Biederman J, Buitelaar J, Castellanos FX, Doyle A, Ebstein RP, et al. Meta-analysis of genome-wide linkage scans of attention deficit hyperactivity disorder. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2008;147B:1392–1398. doi: 10.1002/ajmg.b.30878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Neale BM, Medland SE, Ripke S, Asherson P, Franke B, Lesch KP, Faraone SV, Nguyen TT, Schafer H, Holmans P, et al. Meta-analysis of genome-wide association studies of attention-deficit/hyperactivity disorder. J. Am. Acad. Child Adolesc. Psychiatry. 2010;49:884–897. doi: 10.1016/j.jaac.2010.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Smalley SL, Kustanovich V, Minassian SL, Stone JL, Ogdie MN, McGough JJ, McCracken JT, MacPhie IL, Francks C, Fisher SE, et al. Genetic linkage of attention-deficit/hyperactivity disorder on chromosome 16p13, in a region implicated in autism. Am. J. Hum. Genet. 2002;71:959–963. doi: 10.1086/342732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ogdie MN, Fisher SE, Yang M, Ishii J, Francks C, Loo SK, Cantor RM, McCracken JT, McGough JJ, Smalley SL, et al. Attention deficit hyperactivity disorder: fine mapping supports linkage to 5p13, 6q12, 16p13, and 17p11. Am. J. Hum. Genet. 2004;75:661–668. doi: 10.1086/424387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Poelmans G, Pauls DL, Buitelaar JK, Franke B. Integrated genome-wide association study findings: identification of a neurodevelopmental network for attention deficit hyperactivity disorder. Am. J. Psychiatry. 2011;168:365–377. doi: 10.1176/appi.ajp.2010.10070948. [DOI] [PubMed] [Google Scholar]
- 14.Elia J, Gai X, Xie HM, Perin JC, Geiger E, Glessner JT, D’Arcy M, deBerardinis R, Frackelton E, Kim C, et al. Rare structural variants found in attention-deficit hyperactivity disorder are preferentially associated with neurodevelopmental genes. Mol. Psychiatry. 2010;15:637–646. doi: 10.1038/mp.2009.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Williams NM, Zaharieva I, Martin A, Langley K, Mantripragada K, Fossdal R, Stefansson H, Stefansson K, Magnusson P, Gudmundsson OO, et al. Rare chromosomal deletions and duplications in attention-deficit hyperactivity disorder: a genome-wide analysis. Lancet. 2010;376:1401–1408. doi: 10.1016/S0140-6736(10)61109-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lesch KP, Selch S, Renner TJ, Jacob C, Nguyen TT, Hahn T, Romanos M, Walitza S, Shoichet S, Dempfle A, et al. Genome-wide copy number variation analysis in attention-deficit/hyperactivity disorder: association with neuropeptide Y gene dosage in an extended pedigree. Mol. Psychiatry. 2011;16:491–503. doi: 10.1038/mp.2010.29. [DOI] [PubMed] [Google Scholar]
- 17.Brookes K, Xu X, Chen W, Zhou K, Neale B, Lowe N, Anney R, Franke B, Gill M, Ebstein R, et al. The analysis of 51 genes in DSM-IV combined type attention deficit hyperactivity disorder: association signals in DRD4, DAT1 and 16 other genes. Mol. Psychiatry. 2006;11:934–953. doi: 10.1038/sj.mp.4001869. [DOI] [PubMed] [Google Scholar]
- 18.Johansson S, Halleland H, Halmoy A, Jacobsen KK, Landaas ET, Dramsdahl M, Fasmer OB, Bergsholm P, Lundervold AJ, Gillberg C, et al. Genetic analyses of dopamine related genes in adult ADHD patients suggest an association with the DRD5–microsatellite repeat, but not with DRD4 or SLC6A3 VNTRs. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2008;147B:1470–1475. doi: 10.1002/ajmg.b.30662. [DOI] [PubMed] [Google Scholar]
- 19.Piletz JE, Zhang X, Ranade R, Liu C. Database of genetic studies of bipolar disorder. Psychiatr. Genet. 2011;21:57–68. doi: 10.1097/YPG.0b013e328341a346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Allen NC, Bagade S, McQueen MB, Ioannidis JP, Kavvoura FK, Khoury MJ, Tanzi RE, Bertram L. Systematic meta-analyses and field synopsis of genetic association studies in schizophrenia: the SzGene database. Nat. Genet. 2008;40:827–834. doi: 10.1038/ng.171. [DOI] [PubMed] [Google Scholar]
- 21.Jia P, Sun J, Guo AY, Zhao Z. SZGR: a comprehensive schizophrenia gene resource. Mol. Psychiatry. 2010;15:453–462. doi: 10.1038/mp.2009.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Basu SN, Kollu R, Banerjee-Basu S. AutDB: a gene reference resource for autism research. Nucleic Acids Res. 2009;37:D832–D836. doi: 10.1093/nar/gkn835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Matuszek G, Talebizadeh Z. Autism Genetic Database (AGD): a comprehensive database including autism susceptibility gene-CNVs integrated with known noncoding RNAs and fragile sites. BMC Med. Genet. 2009;10:102. doi: 10.1186/1471-2350-10-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sherry ST, Ward MH, Kholodov M, Baker J, Phan L, Smigielski EM, Sirotkin K. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Flicek P, Amode MR, Barrell D, Beal K, Brent S, Chen Y, Clapham P, Coates G, Fairley S, Fitzgerald S, et al. Ensembl 2011. Nucleic Acids Res. 2011;39:D800–D806. doi: 10.1093/nar/gkq1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Seal RL, Gordon SM, Lush MJ, Wright MW, Bruford EA. genenames.org: the HGNC resources in 2011. Nucleic Acids Res. 2011;39:D514–D519. doi: 10.1093/nar/gkq892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fujita PA, Rhead B, Zweig AS, Hinrichs AS, Karolchik D, Cline MS, Goldman M, Barber GP, Clawson H, Coelho A, et al. The UCSC Genome Browser database: update 2011. Nucleic Acids Res. 2011;39:D876–D882. doi: 10.1093/nar/gkq963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kanehisa M, Goto S, Hattori M, Aoki-Kinoshita KF, Itoh M, Kawashima S, Katayama T, Araki M, Hirakawa M. From genomics to chemical genomics: new developments in KEGG. Nucleic Acids Res. 2006;34:D354–D357. doi: 10.1093/nar/gkj102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lander E, Kruglyak L. Genetic dissection of complex traits: guidelines for interpreting and reporting linkage results. Nat. Genet. 1995;11:241–247. doi: 10.1038/ng1195-241. [DOI] [PubMed] [Google Scholar]
- 31.Coghill D, Banaschewski T. The genetics of attention-deficit/hyperactivity disorder. Expert Rev. Neurother. 2009;9:1547–1565. doi: 10.1586/ern.09.78. [DOI] [PubMed] [Google Scholar]
- 32.Altshuler DM, Gibbs RA, Peltonen L, Dermitzakis E, Schaffner SF, Yu F, Bonnen PE, de Bakker PI, Deloukas P, Gabriel SB, et al. Integrating common and rare genetic variation in diverse human populations. Nature. 2010;467:52–58. doi: 10.1038/nature09298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lasky-Su J, Neale BM, Franke B, Anney RJ, Zhou K, Maller JB, Vasquez AA, Chen W, Asherson P, Buitelaar J, et al. Genome-wide association scan of quantitative traits for attention deficit hyperactivity disorder identifies novel associations and confirms candidate gene associations. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2008;147B:1345–1354. doi: 10.1002/ajmg.b.30867. [DOI] [PubMed] [Google Scholar]
- 34.Lesch KP, Timmesfeld N, Renner TJ, Halperin R, Roser C, Nguyen TT, Craig DW, Romanos J, Heine M, Meyer J, et al. Molecular genetics of adult ADHD: converging evidence from genome-wide association and extended pedigree linkage studies. J. Neural Transm. 2008;115:1573–1585. doi: 10.1007/s00702-008-0119-3. [DOI] [PubMed] [Google Scholar]
- 35.Neale BM, Lasky-Su J, Anney R, Franke B, Zhou K, Maller JB, Vasquez AA, Asherson P, Chen W, Banaschewski T, et al. Genome-wide association scan of attention deficit hyperactivity disorder. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2008;147B:1337–1344. doi: 10.1002/ajmg.b.30866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mick E, Todorov A, Smalley S, Hu X, Loo S, Todd RD, Biederman J, Byrne D, Dechairo B, Guiney A, et al. Family-based genome-wide association scan of attention-deficit/hyperactivity disorder. J. Am. Acad. Child. Adolesc. Psychiatry. 2010;49:898–905 e893. doi: 10.1016/j.jaac.2010.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Neale BM, Medland S, Ripke S, Anney RJ, Asherson P, Buitelaar J, Franke B, Gill M, Kent L, Holmans P, et al. Case-control genome-wide association study of attention-deficit/hyperactivity disorder. J. Am. Acad. Child. Adolesc. Psychiatry. 2010;49:906–920. doi: 10.1016/j.jaac.2010.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. PNAS. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang K, Cui S, Chang S, Zhang L, Wang J. i-GSEA4GWAS: a web server for identification of pathways/gene sets associated with traits by applying an improved gene set enrichment analysis to genome-wide association study. Nucleic Acids Res. 2010;38:W90–W95. doi: 10.1093/nar/gkq324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang K, Chang S, Cui S, Guo L, Zhang L, Wang J. ICSNPathway: identify candidate causal SNPs and pathways from genome-wide association study by one analytical framework. Nucleic Acids Res. 2011;39:W437–W443. doi: 10.1093/nar/gkr391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mailman MD, Feolo M, Jin Y, Kimura M, Tryka K, Bagoutdinov R, Hao L, Kiang A, Paschall J, Phan L, et al. The NCBI dbGaP database of genotypes and phenotypes. Nat. Genet. 2007;39:1181–1186. doi: 10.1038/ng1007-1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liberzon A, Subramanian A, Pinchback R, Thorvaldsdottir H, Tamayo P, Mesirov JP. Molecular signatures database (MSigDB) 3.0. Bioinformatics. 2011;27:1739–1740. doi: 10.1093/bioinformatics/btr260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ferreira MA, O'Donovan MC, Meng YA, Jones IR, Ruderfer DM, Jones L, Fan J, Kirov G, Perlis RH, Green EK, et al. Collaborative genome-wide association analysis supports a role for ANK3 and CACNA1C in bipolar disorder. Nat. Genet. 2008;40:1056–1058. doi: 10.1038/ng.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Askland K, Read C, Moore J. Pathways-based analyses of whole-genome association study data in bipolar disorder reveal genes mediating ion channel activity and synaptic neurotransmission. Hum. Genet. 2009;125:63–79. doi: 10.1007/s00439-008-0600-y. [DOI] [PubMed] [Google Scholar]
- 45.Pal P, Mihanovic M, Molnar S, Xi H, Sun G, Guha S, Jeran N, Tomljenovic A, Malnar A, Missoni S, et al. Association of tagging single nucleotide polymorphisms on 8 candidate genes in dopaminergic pathway with schizophrenia in Croatian population. Croat. Med. J. 2009;50:361–369. doi: 10.3325/cmj.2009.50.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cubells JF, Sun X, Li W, Bonsall RW, McGrath JA, Avramopoulos D, Lasseter VK, Wolyniec PS, Tang YL, Mercer K, et al. Linkage analysis of plasma dopamine beta-hydroxylase activity in families of patients with schizophrenia. Hum. Genet. 2011;130:635–643. doi: 10.1007/s00439-011-0989-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Punia S, Das M, Behari M, Mishra BK, Sahani AK, Govindappa ST, Jayaram S, Muthane UB, K TB, Juyal RC. Role of polymorphisms in dopamine synthesis and metabolism genes and association of DBH haplotypes with Parkinson's disease among North Indians. Pharmacogenet. Genomics. 2010;20:435–441. doi: 10.1097/FPC.0b013e32833ad3bb. [DOI] [PubMed] [Google Scholar]
- 48.Stein LD, Mungall C, Shu S, Caudy M, Mangone M, Day A, Nickerson E, Stajich JE, Harris TW, Arva A, et al. The generic genome browser: a building block for a model organism system database. Genome Res. 2002;12:1599–1610. doi: 10.1101/gr.403602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stergiakouli E, Thapar A. Fitting the pieces together: current research on the genetic basis of attention-deficit/hyperactivity disorder (ADHD) Neuropsychiatr. Dis. Treat. 2010;6:551–560. doi: 10.2147/NDT.S11322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang K, Li M, Hakonarson H. Analysing biological pathways in genome-wide association studies. Nat. Rev. Genet. 2010;11:843–854. doi: 10.1038/nrg2884. [DOI] [PubMed] [Google Scholar]