Abstract
The Minimum Information Required in the Annotation of Models Registry (http://www.ebi.ac.uk/miriam) provides unique, perennial and location-independent identifiers for data used in the biomedical domain. At its core is a shared catalogue of data collections, for each of which an individual namespace is created, and extensive metadata recorded. This namespace allows the generation of Uniform Resource Identifiers (URIs) to uniquely identify any record in a collection. Moreover, various services are provided to facilitate the creation and resolution of the identifiers. Since its launch in 2005, the system has evolved in terms of the structure of the identifiers provided, the software infrastructure, the number of data collections recorded, as well as the scope of the Registry itself. We describe here the new parallel identification scheme and the updated supporting software infrastructure. We also introduce the new Identifiers.org service (http://identifiers.org) that is built upon the information stored in the Registry and which provides directly resolvable identifiers, in the form of Uniform Resource Locators (URLs). The flexibility of the identification scheme and resolving system allows its use in many different fields, where unambiguous and perennial identification of data entities are necessary.
INTRODUCTION
The size and complexity of data produced in biology has made it increasingly important to provide metadata alongside the core data itself. This metadata may comprise domain-specific information as described by minimal information ‘checklists’ meant to enable accurate data reuse or may be ontological in nature, specifying more precisely the kind of entities under consideration. Community-level collaborative bodies such as Minimum Information for Biological and Biomedical Investigations (MIBBI) (1) and Open Biomedical Ontologies (OBO) (2) exist to formalize and coordinate such efforts across the Life Sciences. The computational systems biology community developed one such checklist, entitled the Minimum Information Required in the Annotation of Models (MIRIAM) (3), in order to define the meta-information needed to ensure the re-usability of computational models of biological processes. These guidelines describe the need to unambiguously and perennially identify model components, as well as other information regarding model origin and development. When used in conjunction with standard computer-readable formats such as Systems Biology Markup Language (SBML) (4), controlled annotations facilitate not only model reuse, but also permit efficient search strategies, accurate model comparison and meaningful model conversion between different formats. Furthermore, the relevant linking of models to biological knowledge transforms them into repositories of information.
In order to provide globally unique, perennial and location-independent identifiers for data used in the biomedical domain, we developed MIRIAM Identifiers and the MIRIAM Registry (5). MIRIAM Identifiers are Uniform Resource Identifiers (URIs), which unambiguously identify a record in a data collection, independently of the specific resources distributing instances of those records. The MIRIAM Registry provides information about the different data collections and how to access instances of their records. Definitions of the terms used in subsequent descriptions are detailed in the Table 1 (definition).
Table 1.
Definitions
| Data collection | A data collection gathers data of the same type (e.g. DNA, RNA or protein) and stores information regarding the same sets of ‘properties’ (e.g. sequence, references). It should make use of a well-defined internal identifier scheme. For example, the namespace ‘uniprot’ identifies a data collection whose subject is proteins, whose representation is protein sequence-centric, where each entry stores protein domain information and where the identifier scheme can be described using a specific regular expression. Similarly, ‘ec-code’ identifies a data collection that provides access to enzyme records and ‘chebi’ to an ontological representation of chemicals. |
| Resource | A resource is the physical location on the Web where information about a data record can be accessed. A resource provides the instances of all the records belonging to a collection. Since the record identifier is independent of physical location (URL), it may be resolved using any of the resources listed for that data collection. |
| Namespace | The namespace is the unique syntactic string which defines a data collection. For example, given the identifier ‘urn:miriam:ec-code:1.1.1.1’, the namespace is defined as ‘ec-code'. This precise lexical string is used in both URN and URL forms of the identifiers. |
MIRIAM IDENTIFIERS
To completely fulfil its roles, an identifier must be: (i) unique (two identifiers should not be associated with the same entity); (ii) unambiguous (an identifier must only be associated with a single entity); (iii) perennial (the same identifier should remain associated with an entity for the whole duration of its existence). In addition, an identifier should preferably also be (iv) standard compliant (for easier software support); (v) resolvable (convertible into a physical address on the World Wide Web); and (vi) free to use. MIRIAM identifiers were designed to satisfy all these criteria (5).
In order to provide a unique identifier for a record, regardless of the physical location(s) where that information can be retrieved, MIRIAM identifiers are composed of three parts. The first part is a prefix, dependent on the scheme used (see below) and that specifies ‘this is a MIRIAM URI’. The second part is the namespace that identifies the data collection. The third and final part is the internal identifier of a specific record in a data collection (this identifier is created and provided by the data collection itself). MIRIAM URIs were initially only provided as Uniform Resource names (URNs). The prefix of the URN scheme is urn:miriam. For example, the enzyme alcohol dehydrogenase in the enzyme classification collection is identified by urn:miriam:ec-code:1.1.1.1 and the species Homo sapiens in the taxonomy of living species by urn:miriam:taxonomy:9606.
In order to access the data for a record, one must rely on a URI resolving system, such as the MIRIAM Registry described below. The URNs can be resolved into URLs, for instance, using Web Services or by processing the XML export of the MIRIAM Registry. But they are not directly resolvable, so one cannot successfully copy/paste them into a browser and get an informative page. In order to provide directly dereferencable URIs and comply with the second rule of Linked Data (6), we recently introduced a new URL-based identification scheme. This scheme provides directly resolvable identifiers, based on the information stored in the MIRIAM Registry. The prefix of the URL scheme is http://identifiers.org/. This identification scheme runs entirely in parallel with the URN form of MIRIAM identifiers. Both forms essentially share the same structure, are based on the same shared list of namespaces and are fully inter-convertible. For example, the enzyme alcohol dehydrogenase in the enzyme classification collection is identified by http://identifiers.org/ec-code/1.1.1.1 and the species Homo sapiens in the taxonomy of living species by http://identifiers.org/taxonomy/9606.
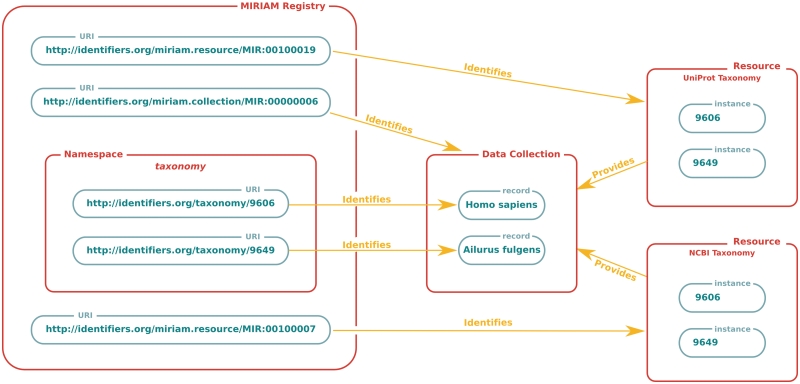
The key difference between the two parallel schemes, which offer exactly the same access to data, is that the Identifiers.org URLs can be resolved directly and do not require special software tools on the user side for their handling; the dereferencing is performed by the Identifiers.org service (see below). The inter-relationships between the information captured by this service are illustrated in Figure 1.
Figure 1.
Concepts and component information captured in the MIRIAM Registry. The MIRIAM Registry collects information about data collections and resources, allowing them to be referenced using URIs. Red-bounded boxes represent concepts, while green ones depict specific instances. Each collection, which itself can be referenced via a URI, is assigned a namespace. This namespace can be combined with a suitable identifier in order to form a URI identifying the specific data record, independently of any physical locations holding that information. Each of these resolvable physical locations are regarded as an instance of the data record, and can themselves be identified using a URI.
MIRIAM REGISTRY
While the location of information on the Web is a convenient endpoint for cross-references, it is fraught with issues which can result in ‘dead links'. These can be caused by changes in the underlying infrastructure of a resource or in modification of the access URL or identification scheme used by that resource. In addition, data of a given collection can often be accessed via several providers on the Web, for example, when it is ‘mirrored’ but also when associated with different metadata. In these cases, the record is identical (the data relevant to the collection describing the entity), though the physical instances of the record differ. MIRIAM Registry tackles this problem through the recording of resources, which are physical locations (associated with URLs) where one can access the entity or information about the entity. The concept of resource effectively allows the decoupling of the identification of an entity from its location on the Web, enabling the association of a single entity identifier with multiple locations. Resources and data collections are themselves identified as records in the MIRIAM Registry and have their own namespace. For instance, the enzyme nomenclature is identified by http://identifiers.org/miriam.collection/MIR:00000004 while the enzyme nomenclature distributed by the Swiss Institute of Bioinformatics is http://identifiers.org/miriam.resource/MIR:00100003. For a detailed list of all the information stored for each data collection, refer to Table 2 (information).
Table 2.
Information
| Data collection information | |
| Identifier | A stable MIRIAM Registry identifier of the data collection. |
| Name | The name usually used to refer to the data collection. |
| Synonym(s) | Alternative name(s) of the data collection. |
| Namespace | The part of the URIs which identifies the data collection. For example ‘ec-code’ for enzymes. |
| Deprecated root URI(s) | MIRIAM URNs or URLs that have become obsolete over time. Deprecated identifiers are stored in the Registry, allowing conversion to current forms. |
| Definition | Short description of the data collection, indicating the focus of its content. |
| Identifier pattern | A regular expression pattern that describes the identifiers used within the data collection. |
| Reference(s) | Link(s) to documentation about the data collection and relevant publication(s). |
| Resource information | |
| Identifier | Each resource associated with a collection is given a unique identifier in the MIRIAM Registry. |
| Access URL | URL used to retrieve a given data entry, where the token ($id) is replaced with a specified identifier for a record. |
| Website | Root URL of the resource, usually its home page. |
| Description | Brief description about the resource, used to distinguish the current resource from all the others recorded for the same data collection. |
| Institution | The institution responsible for hosting the resource. |
| Health status | Though not a textual field, the resource health status is displayed by the colour-coded text area. |
| Deprecated Physical Location(s) | A list of deprecated resource(s) which are no longer usable to resolve information for this data collection. |
Initial population of the collections in the MIRIAM Registry came largely from the Systems Biology community, and more specifically from those collections that were commonly used in the annotation of models stored in BioModels Database (7). Further collections have been incorporated based on the requests from individual users, from collaborative bodies, such as the Protein Standards Initiative (http://www.psidev.info/) and from publicly available listings of Life Sciences databases, such as those used in cross-referencing (for example, http://www.geneontology.org/cgi-bin/xrefs.cgi). The public-facing web application provides easy access to the catalogue of data collections. Any visitor to the site can suggest modification of the information recorded (through a link at the bottom of each page) and submit new data collections (through a dedicated form linked from the left menu panel). More detailed information on making submissions and ascertaining the suitability of a collection for inclusion can be found here in the FAQ (http://www.ebi.ac.uk/miriam/main/mdb?section=faq).
All records are manually curated in order to ensure accuracy and consistency. When the information provided by the submitter, usually obtained from the relevant resources on the web, is incomplete, we liaise with the developers and administrators of those collections. This is particularly important when issues arise, for example, regarding the identifier schemes used by the collection or the specific details of the license under which the information is made available.
The MIRIAM Registry infrastructure is written in Java (http://java.sun.com/javaee/) and makes use of the Model-View-Controller design pattern (http://en.wikipedia.org/wiki/Model-view-controller). The application runs inside an Apache Tomcat Web container (http://tomcat.apache.org/). All the information in the Registry is stored in a MySQL database (http://www.mysql.com/).
Since its original introduction, there has been significant growth and development of the MIRIAM Registry. It currently contains information on over 250 collections, with a further 64 undergoing the curation process. The Registry also provides supporting facilities to enable the convenient usage of both URN and URL identification schemes.
Finding collections and resources
With the plethora of available data collections potentially suitable to annotate biomedical information, it can be somewhat problematic to locate the most appropriate one. To aid in this task, each collection is associated with one or more tags. For instance, should a user require a data collection with which to annotate a protein sequence, it is possible to search using the two tags ‘protein’ and ‘sequence’ to find suitable data collections. This ‘Tags’ search function is linked from the left menu panel and displays a selectable list of the tags used. There are currently 39 tags available with which collections can be associated. These were created in ad hoc fashion when the system was implemented and currently are sufficient in number to allow the existing collections to be associated with two to three tags each. Additional tags can be created as needed by curators or as requested by users. The tags are of a coarse granularity, describing the type of data recorded (‘sequence’ ‘expression’ ‘phenotype’), the subject of those data (‘gene’ ‘protein’ ‘drug’), the domain area to which they relate (‘disease’ ‘pharmacogenomics’ ‘neuroscience’) or taxonomic associations of the data (‘mammalian’ ‘human’). The purpose of the tagging system is to allow users to identify appropriate collections based on a gross level query. Plans to improve this tagging mechanism, for instance by incorporating ontological information at the level of resources and the data they provide, are discussed below.
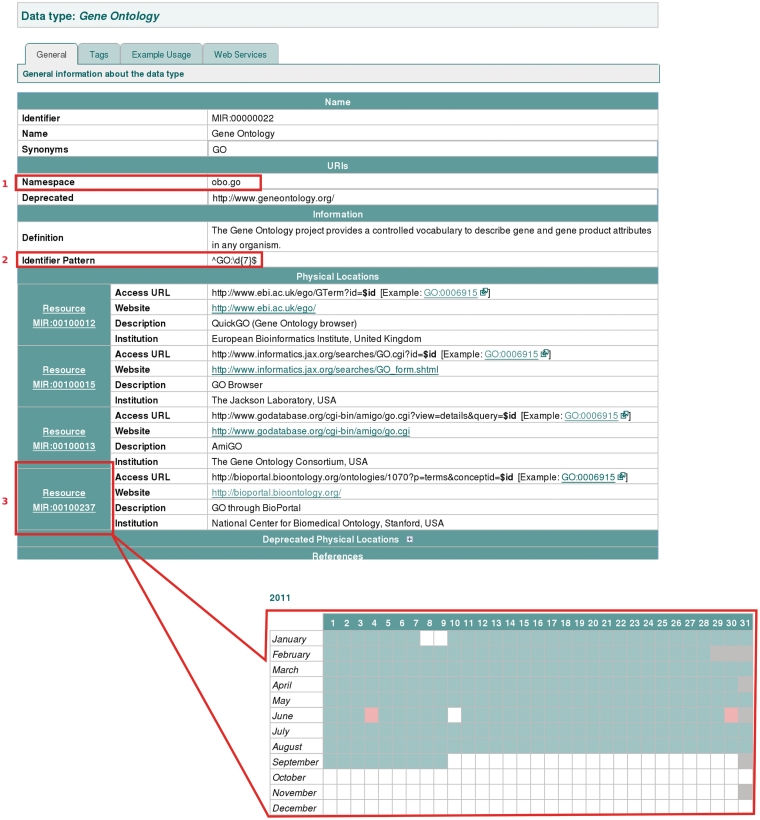
The MIRIAM Registry provides access to several resources serving the same data collection. Those resources are not necessarily identical and there may be reasons to prefer one over another. To allow users to make an informed choice when selecting such a resource, we provide additional information, including the uptime of the servers running the service. For this purpose a ‘health status’ has been implemented and a daily health check is automatically performed for each resource listed in the Registry (Figure 2). A summary of the health status of a specific resource is depicted by colour coding where green indicates an uptime in excess of 90% and graduated colour coding with downtime to below 20% being represented in red. More detailed information is available (by clicking on a resource's identifier), such as a calendar view of the uptime and details of the last check made. The system is also used by the Registry curators as a warning system to highlight otherwise unnoticed changes in the way data is accessed from a particular resource.
Figure 2.
An illustration of the variety of information captured for each data collection in the MIRIAM Registry. Some fields, described in the Information Table 2, are highlighted: (1) Namespace; (2) Identifier pattern, which allows automated checking of identifier validity with respect to the expected expression pattern; and (3) Resource health status, which provides information on resource up- and down-time. A notification of the health status is given through colour coding, while more details are presented on a separate page, via a link on the resource identifier (see inset).
Programmatic use of the Registry
Simple Object Access Protocol (SOAP) and REpresentational State Transfer (REST) access methods are available to query the Registry. These Application Programming Interfaces (APIs) can be used to generate and resolve the identifiers, as well as to extract information about individual resources providing access to the data records.
To facilitate the usage of the web services by third party tools, a Java library is provided. It allows querying of the Registry in a quick and convenient way. It is available for download from the SourceForge.net project (http://sourceforge.net/projects/miriam/).
The entire content of the Registry is also made available as an XML file export. This file is auto-generated daily and additionally can be created on demand.
IDENTIFIERS.ORG RESOLVING SYSTEM
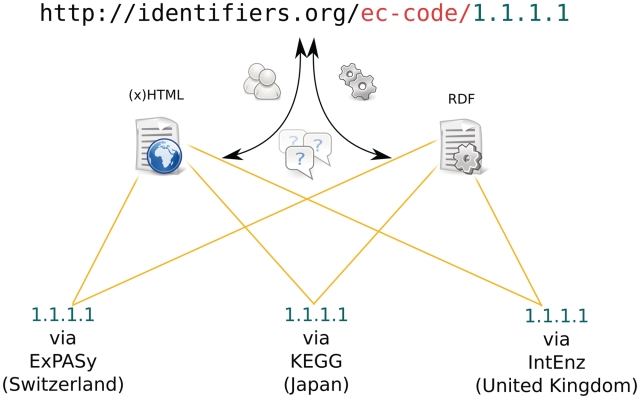
Identifiers.org is the resolving system of MIRIAM identifiers’ URL form. For more information, readers may refer to http://identifiers.org/. Access to a given collection in the Registry, such as the enzyme nomenclature is given by appending the namespace, as in http://identifiers.org/ec-code/. Due to the decoupling between the data collections and the resources that provide record information, the resolution of an Identifiers.org URL, such as http://identifiers.org/ec-code/1.1.1.1, directs the user to an intermediate page listing all recorded physical locations where a record may be accessed, allowing the user to choose the most suitable one. This process is illustrated in Figure 3.
Figure 3.
An illustration of the process followed when dereferencing an Identifiers.org URL. The example URL is a location-independent identifier for an ec-code record. When used in a browser, it resolves to an intermediate HTML page that provides a list of possible physical locations where the data record can be retrieved. The default format of this document is HTML, while an RDF/XML version is available via content negotiation or by using the ‘format’ parameter in the URL (see the ‘Identifiers.org Resolving System’ section).
One can directly access the instance of a record in a given resource by appending a parameter as a suffix. For instance, the following URL provides access to the record for alcohol dehydrogenase of the enzyme nomenclature collection provided by the IntEnz resource: http://identifiers.org/ec-code/1.1.1.1?resource=MIR:00100001. Alternatively, the concept of ‘profile’ allows one to customize the behaviour of the resolving system: it allows the pre-selection of resources to be used in the dereferencing, for a whole range of data collections. For example, the pre-defined profile ‘most_reliable’ as in http://identifiers.org/ec-code/1.1.1.1?profile=most_reliable, always returns the instance of a record in the resource with the best uptime. The ‘most_reliable’ profile is currently based on the health check history over the whole lifetime of the resource since its inclusion in the Registry. The valid parameters available for use are illustrated at: http://identifiers.org/examples/.
The information about all the instances of a record is presented by default as HTML, but may also be retrieved in RDF/XML format. Either format can be recovered through content negotiation or using the ‘format’ parameter within the URL (for example: http://identifiers.org/ec-code/1.1.1.1?format=rdfxml). It is possible to accommodate further output formats as requested by the user community. The information represented in an RDF form allows the additional incorporation of semantic information. These semantics are captured using standard vocabularies such as SIO (Semanticscience Integrated Ontology; http://semanticscience.org/ontology/sio.owl) and EDAM (EMBRACE Data and Methods; http://edamontology.sourceforge.net/), using terms such as ‘has_identifier’ and ‘accession’ to describe relationships and data concepts, respectively.
Parameter specification should be used only in conjunction with user interface instantiation (for example when used in a browser) and should be avoided when a URL is to be used as unique identifier for a collection record. There are a number of potential parameter name–value combinations possible for URIs, making a direct comparison difficult if provided with accompanying parametrization. Hence, in the unambiguous and perennial identification of data, the ‘atomic’ identifier should be considered as the minimal string that specifies the record.
The resolving system also provides direct information for malformed queries, for example, where the identifier is not properly encoded, or when a deprecated URL is used. In both cases a clear message is given to inform users of the situation. In addition to the human-readable description of the error, the system also returns the appropriate HTTP status code.
CURRENT STATUS AND FUTURE DEVELOPMENTS
MIRIAM URNs are already widely used, particularly within the Computational Systems Biology community. For example, in the 20th release (September 2011) of BioModels Database, the 764 models contain over 25 000 MIRIAM identifiers.
An overview of the widespread use of MIRIAM Registry information and its identifiers is detailed on the website (http://www.ebi.ac.uk/miriam/main/mdb?section=use). URI identifiers are supported by a variety of file formats; software libraries and tools have been developed to generate, resolve and leverage upon MIRIAM URIs in novel scientific research. Registry namespaces are being used as controlled vocabularies in databases and standardization efforts. Moreover, the Life Science Registry Name identification scheme, which will imminently no longer be developed and supported, has decided upon Identifiers.org as its replacement and successor.
The launch of the Identifiers.org URL as an alternative to the URN form of MIRIAM identifiers answers the ever-growing need to provide directly resolvable identifiers, especially for Semantic Web applications, such as the Linking Open Data initiative from the W3C. Collections and resources used in those efforts, such as those from the Life Science Dataset Registry which supports the Bio2RDF project (8), are currently being integrated in the Registry.
To enable the incorporation of a wider array of data collections, the Registry is being updated to store additional information, such as restrictions on the access to the data entries (for example the need to register and login), on its use (utilization of specific license and/or copyright), etc. In addition, resource and collection descriptions will be further enhanced by the incorporation of information from ontologies such as the Biomedical Resource Ontology (http://bioportal.bioontology.org/ontologies/1104). These modifications will allow users to ascertain the appropriateness of use of particular data collections and will improve existing search facilities.
Profiles predefine specific resolving locations for each selected data collection and may be shared using the ‘profile’ parameter described above. Moreover, there is currently ongoing work to allow users to create profiles in the Registry through a dedicated user interface. This interface will also list the publicly available profiles that have been created by users, stating the collection and preferred resources associated.
The MIRIAM Registry is a collaborator of the BioDBCore effort (9). This effort focuses on a community approved minimum information checklist to which database providers should comply. It recommends that the standards that are implemented by a data provider (such as which standard formats it accepts and provides, which terminologies it uses, etc.) should be recorded with reference to those standards listed by BioSharing (http://biosharing.org). This information, ideally, would be provided by database administrators as an RDF file. The BioDBCore database will rely on dedicated resources for the storage of some information. Identifiers.org URIs will be used for identification and data access information.
All the code used to develop the MIRIAM Registry and the associated helper utilities are released under the terms of the GNU, General Public License and are available at: http://sourceforge.net/projects/miriam/.
CONCLUSIONS
The MIRIAM Registry is a stable resource, which provides both an identifier scheme and resolution system. While it originates from within the Computational Systems Biology community, it is certainly not limited to that domain, submissions are encouraged for new data collections from any biological community that has a desire to create and/or use unambiguous perennial identifiers or to generate and resolve physical locations from which data can be accessed. The system is of particular interest to tool and database developers who need to manage annotations and cross-references. Identifiers.org helps to ensure that data entities are resolvable, thereby avoiding the creation of ‘dead ends’ in the network of linked data.
FUNDING
EMBL, ELIXIR (Preparatory Phase) and BBSRC (grants BB/E005748/1 and JPA 1729). Funding for open access charge: core EMBL funding.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors thank Michel Dumontier for very fruitful discussions about identifiers and their usage in the Semantic Web, as well as existing users for their participation and feedback in discussions and through surveys.
REFERENCES
- 1.Taylor CF, Field D, Sansone S-A, Aerts J, Apweiler R, Ashburner M, Ball CA, Binz PA, Bogue M, Booth T, et al. Promoting coherent minimum reporting guidelines for biological and biomedical investigations: the MIBBI project. Nat. Biotechnol. 2008;26:889–896. doi: 10.1038/nbt.1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Smith B, Ashburner M, Rosse C, Bard J, Bug W, Ceusters W, Goldberg LJ, Eilbeck K, Ireland A, Mungall CJ, The OBI Consortium, Leontis,N., Rocca-Serra,P., Ruttenberg,A., Sansone,S.-A., Scheuermann,R.H., Shah,N., Whetzel,P.L., Lewis,S The OBO Foundry: coordinated evolution of ontologies to support biomedical data integration. Nat. Biotechnol. 2007;25:1251–1255. doi: 10.1038/nbt1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Le Novère N, Finney A, Hucka M, Bhalla US, Campagne F, Collado-Vides J, Crampin EJ, Halstead M, Klipp E, Mendes P, et al. Minimum information requested in the annotation of biochemical models (MIRIAM) Nat. Biotechnol. 2005;23:1509–1515. doi: 10.1038/nbt1156. [DOI] [PubMed] [Google Scholar]
- 4.Hucka M, Finney A, Sauro H, Bolouri H, Doyle J, Kitano H, Arkin A, Bornstein B, Bray D, Cuellar A, et al. The Systems Biology Markup Language (SBML): a medium for representation and exchange of biochemical network models. Bioinformatics. 2003;19:524–531. doi: 10.1093/bioinformatics/btg015. [DOI] [PubMed] [Google Scholar]
- 5.Laibe C, Le Novère N. MIRIAM Resources: tools to generate and resolve robust cross-references in Systems Biology. BMC Syst. Biol. 2007;1:58–66. doi: 10.1186/1752-0509-1-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berners-Lee T. 2006. Linked Data, In Design Issues: Architectural and Philosophical Points. http://www.w3.org/DesignIssues/LinkedData (13 September 2011, date last accessed) [Google Scholar]
- 7.Li C, Donizelli M, Rodriguez N, Dharuri H, Endler L, Chelliah V, Li L, He E, Henry A, Stefan MI, et al. BioModels Database: An enhanced, curated and annotated resource for published quantitative kinetic models. BMC Syst. Biol. 2010;4:92. doi: 10.1186/1752-0509-4-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Belleau F, Nolin M, Tourigny N, Rigault P, Morissette J. Bio2RDF: towards a mashup to build bioinformatics knowledge systems. J. Biomed. Inform. 2008;41:706–716. doi: 10.1016/j.jbi.2008.03.004. [DOI] [PubMed] [Google Scholar]
- 9.Gaudet P, Bairoch A, Field D, Sansone S-A, Taylor C, Attwood TK, Bateman A, Blake JA, Bult CJ, Cherry JM, et al. Towards BioDBcore: a community-defined information specification for biological databases. Nucleic Acids Res. 2011;39:D7–D10. doi: 10.1093/nar/gkq1173. [DOI] [PMC free article] [PubMed] [Google Scholar]