Abstract
In addition to maintaining the GenBank® nucleic acid sequence database, the National Center for Biotechnology Information (NCBI) provides analysis and retrieval resources for the data in GenBank and other biological data made available through the NCBI Website. NCBI resources include Entrez, the Entrez Programming Utilities, MyNCBI, PubMed, PubMed Central (PMC), Gene, the NCBI Taxonomy Browser, BLAST, BLAST Link (BLink), Primer-BLAST, COBALT, Splign, RefSeq, UniGene, HomoloGene, ProtEST, dbMHC, dbSNP, dbVar, Epigenomics, Genome and related tools, the Map Viewer, Model Maker, Evidence Viewer, Trace Archive, Sequence Read Archive, BioProject, BioSample, Retroviral Genotyping Tools, HIV-1/Human Protein Interaction Database, Gene Expression Omnibus (GEO), Probe, Online Mendelian Inheritance in Animals (OMIA), the Molecular Modeling Database (MMDB), the Conserved Domain Database (CDD), the Conserved Domain Architecture Retrieval Tool (CDART), Biosystems, Protein Clusters and the PubChem suite of small molecule databases. Augmenting many of the Web applications are custom implementations of the BLAST program optimized to search specialized data sets. All of these resources can be accessed through the NCBI home page at www.ncbi.nlm.nih.gov.
INTRODUCTION
The National Center for Biotechnology Information (NCBI) at the National Institutes of Health was created in 1988 to develop information systems for molecular biology. In addition to maintaining the GenBank® (1) nucleic acid sequence database, which receives data through the international collaboration with DDBJ and EMBL-Bank as well as from the scientific community, NCBI provides data retrieval systems and computational resources for the analysis of GenBank data and many other kinds of biological data. For the purposes of this article, after a summary of recent developments and an introduction to the Entrez system, the NCBI suite of resources is grouped into 10 broad categories based on those in the NCBI guide. All resources discussed are available from the NCBI guide at www.ncbi.nlm.nih.gov and can also be located using the Site Search database. In most cases, the data underlying these resources and executables for the software described are available for download at ftp.ncbi.nih.gov.
RECENT DEVELOPMENTS
BioProject
The BioProject database (www.ncbi.nlm.nih.gov/bioproject/) is a redesigned and expanded replacement of the NCBI Genome Project resource. BioProject enables users to submit comprehensive research studies ranging from focused genome sequencing projects to large international collaborations with multiple subprojects incorporating experiments resulting in nucleotide sequence sets, genotype/phenotype data, sequence variants, or epigenetic information. BioProject also allows users to search for and retrieve data sets that are often difficult to find due to inconsistent annotation, multiple independent submissions and the varied nature of diverse data types that are often stored in different databases. The Limits page provides numerous ways to filter the various project types; for example, checking the ‘Organism overview’ box and searching with the term ‘human[orgn]’ provides a single record summarizing the available projects for the human genome.
BioSample
The BioSample database (www.ncbi.nlm.nih.gov/biosample/) is a new resource that provides annotation for biological samples used in a variety of studies submitted to NCBI, including genomic sequencing, microarrays, GWAS and epigenomics (2). The primary aim of BioSamples is to surmount annotation inconsistencies between similar samples from different studies so that investigators can more easily make connections between all of the available data for a particular sample. Currently BioSample contains over 600 000 samples, with 90% of these coming from either short-read archive (SRA) or dbGaP.
CloneDB and clone finder
NCBI has replaced the Clone Registry with the new CloneDB (www.ncbi.nlm.nih.gov/clone/), a resource for finding descriptions, sources and detailed statistics on available genomic libraries and clones from genome-sequencing projects. The new Library Browser allows filtering by organism, vector type, distributors and number of associated end or insert sequences. The linked Clone Finder quickly identifies clones from these libraries that correspond to regions on assembled genomes, and locates these clones by chromosomal position or by features such as genes, SNPs, markers or transcript sequence accession numbers. The graphical display in Clone Finder shows features annotated on the genome including assembled contigs, their components, genes and aligned transcripts.
Cn3D 4.3 and MMDB updates
NCBI recently released Cn3D 4.3 (www.ncbi.nlm.nih.gov/Structure/CN3D/cn3d.shtml), the newest version of the popular software package for viewing 3D molecular structures and alignments. New features include stereo views, new alignment algorithms, more powerful highlighting features and full communication with CDTree, a separate application for manipulating Conserved Domain alignments. In addition, the MMDB record pages now display clearer depictions of the biological and asymmetric units from the source PDB files, and provide links to load these views into Cn3D 4.3. These pages also contain a new table detailing the molecular interactions occurring within the structure, for example between protein chains and ligands.
PopSet redesign
The PopSet database (www.ncbi.nlm.nih.gov/popset/) is a collection of related sequences and alignments derived from population, phylogenetic, mutation and ecosystem studies that have been submitted to GenBank. In the past year, NCBI completely redesigned the web interface for this database, adding several new features including an embedded graphical alignment viewer and improved integration of data from other PopSets and NCBI databases. For PopSets with fewer than 100 sequences, links are provided to generate a BLAST alignment of the sequences or, if an alignment was submitted as part of the record, a distance tree view of the alignment.
Updates to the Database of Short Genetic Variations (dbSNP)
The Database of Short Genetic Variations (dbSNP) was originally created to support large scale polymorphism discovery (e.g. HapMap), but was rapidly adopted by the scientific community as the world archive for additional classes of variations such as insertions/deletions, microsatellites and non-polymorphic variants. This broadening of the scope of the database has made the dbSNP name misleadingly restrictive, and has led to confusion by some groups wishing to use dbSNP as a useful surrogate for ‘known polymorphisms’ in calling clinical mutations. NCBI is addressing this problem by retaining the traditional dbSNP acronym, but changing the title of the database to ‘short genetic variations’. In addition, we are highlighting the attributes about the clinical or polymorphic character of SNPs on the website, and have added several new SNP attributes: allele origin (germline, somatic or unknown), clinical significance, global minor allele frequency (MAF) and suspect (false positives) (www.ncbi.nlm.nih.gov/projects/SNP/docs/rs_attributes.html). Using these attributes, we are preparing specially filtered variant call format data files to provide reasonable subsets of dbSNP selected to act as surrogates for ‘polymorphic’ or ‘clinically significant’ variants. These subsets are defined using MAFs and sources, and the individual files include the following: (i) MAF > 0.01 based on 1000 Genomes populations; (ii) MAF > 0.01 for any population submitted to dbSNP; (iii) MAF > 0.01 for all populations submitted to dbSNP; (iv) variations asserted to be clinically significant by submitters; and (v) variations suspected to be false positives.
BLAST updates
The genomic and SNP BLAST pages, both accessible from the main BLAST page (blast.ncbi.nlm.nih.gov), have now been updated to the standard BLAST search form used for blastn and blastp searches. These new forms make many additional BLAST options available to users. For example, users can optimize a search by selecting different algorithms in the ‘Program Selection’ section, can exclude model sequences and sequences from uncultured or environmental samples (where applicable), limit a search by an Entrez query and assign a title to a search. Additionally the forms provide only those database options relevant to the current search. The BLAST reports allow users to save parameter sets in MyNCBI for future use and to submit a modified search using the ‘Edit and Resubmit’ link near the top of each report. The genomic BLAST pages can search genomic DNA along with other data sets, such as RefSeq protein annotations, from over 120 organisms available in the NCBI Map Viewer. The SNP page uses RefSNP flanking sequences as the source for its sequence databases.
The SRA BLAST page, accessible from the ‘Specialized BLAST’ section of the main BLAST page, can search Whole-Genome Shotgun (WGS) and transcript sequences from 454 sequencing systems. The SRA sequences are grouped by genus in a pull-down menu, and if multiple species have data within a genus, a separate menu appears allowing individual species to be selected. These data sets are updated daily, so new SRA data is available for searching quickly. The SRA BLAST reports have standard features like ‘edit and resubmit’ and a distance tree presentation.
NCBI also introduced two new BLAST services in the past year. The first is RefSeqGene BLAST, a specialized search of the RefSeqGene collection, described below. The second is simple object access protocol (SOAP)-BLAST, a web service using the SOAP protocol for submitting searches and retrieving results from the NCBI BLAST servers. Additional information about SOAP-BLAST is available at www.ncbi.nlm.nih.gov/books/NBK55699/.
MyNCBI updates
MyNCBI provides users with a wide range of services such as saving search queries, setting up automatic searches with e-mail alerts, storing and organizing NCBI database records, selecting preferred display formats, choosing filtering options and tracking recent usage history. In the past year, the MyNCBI user interface has undergone a comprehensive redesign with an emphasis on immediate access to data and user customization controls. A user's MyNCBI homepage now presents searches, filters, collections and bibliographic entries immediately upon signing in and each tool can be rearranged on the screen or dismissed from the view. The MyNCBI home page also includes two new tools that allow users to track their recent BLAST activity and to launch a text search in any of the NCBI databases. My Bibliography, which can store a wide variety of citations and track compliance with the NIH Public Access Policy, has been enhanced to display related citations, citing articles in PubMed Central (PMC) and links to free full text where applicable.
Updates to literature resources
In the past year, NCBI released several enhancements to PubMed including reformatted abstract pages to improve readability and the automatic display of highlighted search terms in the search results. PubMed abstracts for PMC articles now display a strip of figures and other images generated from the PMC data. The ‘Send to File’ menu now includes a selection to generate an abbreviated summary citation in a comma separated (CSV) file. Finally, PubMed Mobile (www.ncbi.nlm.nih.gov/m/pubmed/) was released to provide a simplified mobile-friendly web interface for accessing PubMed.
The NLM Catalog and MeSH (Medical Subject Headings) databases were redesigned in the past year to provide a streamlined interface similar to that of PubMed. The NLM Catalog provides bibliographic data for over 1.4 million NLM holdings including journals, books, manuscripts, computer software, audio recordings and other electronic resources. Each record is linked to the NLM LocatorPlus service as well as related catalog records with similar title words or associated MeSH terms. The Journals Database has been retired and journal information is now available in the updated NLM Catalog. The PubMed and NLM Catalog homepages include a link to the ‘Journals in NCBI Databases’ (www.ncbi.nlm.nih.gov/nlmcatalog/journals) page, which provides a limit for NLM Catalog searches to the subset of over 27 000 journals that are referenced in NCBI database records.
THE NCBI GUIDE AND THE ENTREZ SYSTEM
The NCBI guide
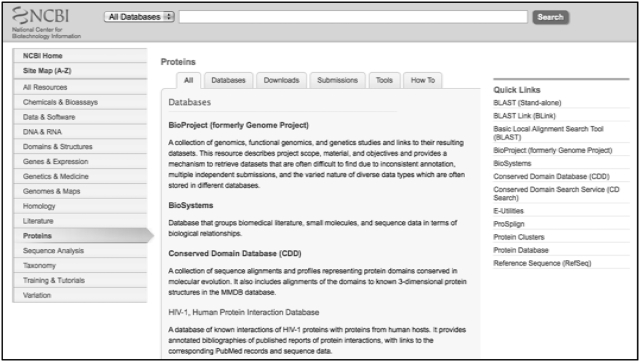
The NCBI guide serves not only as the NCBI home page but also as an interactive directory of the NCBI site. On the main page of the NCBI Guide, the categories in the Resource pull-down menu in the standard header are duplicated in a list on the left of the page. Clicking on any category displays a list of relevant resources sorted into four groups: databases, downloads, submissions and tools (Figure 1). A list of how-to guides is also available via the ‘How-To’ tab on these pages. Popular resources are listed on the right under a ‘Quick Links’ heading, and on the main Guide page, a list of the most frequently used resources is provided in the ‘Popular Resources’ box and also as a list in the standard footer.
Figure 1.
Category page in the NCBI Guide for Proteins. The full list of categories is shown on the left, with Proteins highlighted. The main body of the page displays an alphabetically sorted list of NCBI resources in the Proteins category, and users can further limit this list by clicking on tabs across the top (e.g. protein databases or protein tools). Links to popular resources in the Proteins category are shown under Quick Links on the right.
Entrez databases
Entrez (3) is an integrated database retrieval system that provides access to a diverse set of 35 databases that together contain over 570 million records (Table 1). Entrez supports text searching using simple Boolean queries, downloading of data in various formats and linking of records between databases based on biological relationships. In their simplest form, these links may be cross-references between a sequence and the abstract of the paper in which it is reported or between a protein sequence and its coding DNA sequence or its 3D structure. Computationally derived links between ‘neighboring records’, such as those based on computed similarities among sequences or among PubMed abstracts, allow rapid access to groups of related records. Several popular links are displayed as Discovery Components in the right column of Entrez search result or record view pages, making these connections easier to find and explore. The LinkOut service expands the range of links to include external resources, such as organism-specific genome databases. The records retrieved in Entrez can be displayed in many formats and downloaded singly or in batches.
Table 1.
The Entrez databases (as of September 1, 2011)
| Database | Records | Section within this article | Data sourcea |
|---|---|---|---|
| SNP | 138 764 396 | Genetics and medicine | D (dbSNP), N |
| PubChem substance | 85 365 779 | Chemicals and bioassays | D |
| EST | 70 357 344 | DNA and RNA | D (GenBank) |
| GEO profiles | 63 811 486 | Genes and expression | D |
| Nucleotide | 52 161 410 | DNA and RNA | D (GenBank), C, N |
| Protein | 42 877 630 | Proteins | C, N |
| GSS | 32 578 865 | DNA and RNA | D (GenBank) |
| PubChem compound | 30 455 811 | Chemicals and bioassays | N |
| PubMed | 21 131 087 | Literature | C |
| Probe | 10 386 696 | Genes and expression | D |
| Gene | 8 912 208 | Genes and expression | C, N |
| UniGene | 5 298 131 | Genes and expression | N |
| PMC | 2 257 586 | Literature | D (NIHMS), C |
| NLM catalog | 1 442 076 | Literature | C, N |
| dbVar | 848 633 | Genomes | D |
| Taxonomy | 812 707 | Taxonomy | C, N |
| GEO data sets | 650 717 | Genes and expression | N |
| BioSample | 638 664 | Recent developments | N |
| Protein clusters | 627 757 | Proteins | N |
| UniSTS | 534 208 | Genomes | D (dbSTS) |
| PubChem bioassay | 504 222 | Chemicals and bioassays | D |
| Books | 393 344 | Literature | C, N |
| Biosystems | 257 598 | Genes and expression | C |
| MeSH | 229 533 | Literature | N |
| dbGaP | 138 848 | Genetics and medicine | D |
| PopSet | 136 557 | DNA and RNA | D (GenBank) |
| Homologene | 128 030 | Genes and expression | N |
| SRA | 95 155 | DNA and RNA | D |
| Structure | 75 119 | Domains and structures | C, N |
| CDD | 43 831 | Domains and structures | C, N |
| BioProject | 24 165 | Recent developments | D |
| Genome | 13 846 | Genomes | C, N |
| Site search | 10 640 | Introduction | N |
| Epigenomics | 4241 | Genomes | D |
| OMIA | 2818 | Genetics and medicine | C |
aD = direct submission; C = collaboration/agreement; N = internal NCBI/NLM curation.
Data sources and collaborations
NCBI receives data from three sources; direct submissions from external investigators; collaborations or agreements, both national and international, with data providers and research consortia; and internal curation efforts. The ‘Submissions’ column in Table 1 indicates those mechanisms by which each Entrez database receives data. The various collaborations, agreements and curation efforts are described throughout the remainder of this article.
Entrez programming utilities (E-utilities)
The Entrez Programming Utilities (E-Utilities) constitute the Application Programming Interface (API) for the Entrez system. The API includes eight programs that support a uniform set of parameters used to search, link and download data from the Entrez databases. EInfo provides basic statistics on a given database, including the last update date and lists of all search fields and available links. ESearch returns the identifiers of records that match an Entrez text query, and when combined with EFetch or ESummary, provides a mechanism for downloading the corresponding data records. ELink gives users access to the vast array of links within Entrez so that data related to an input set can be retrieved. By assembling URL or SOAP calls to the E-utilities within simple scripts, users can create powerful applications to automate Entrez functions to accomplish batch tasks that are impractical using web browsers. Instructions for using the E-Utilities are found on the NCBI Bookshelf at www.ncbi.nlm.nih.gov/bookshelf/br.fcgi?book=helpeutils.
LITERATURE
PubMed
The PubMed database now contains more than 21 million citations, some of which date back to the early 1800s, from more than 24 000 life science journals. Over 12 million of these citations have abstracts, and 12.5 million have links to their full text articles, with 9 million having both an abstract and a link to full text. PubMed is heavily linked to other core NCBI databases, thereby providing a crucial bridge between the data of molecular biology and the scientific literature. PubMed records are also linked to one another as ‘related citations’ based on the computationally detected similarities using indexed MeSH (4) terms and the text of titles and abstracts. Succinct descriptions of the top five related citations are shown on the default Abstract display.
PMC
PMC (5) is a digital archive of peer-reviewed journal articles in the life sciences and now contains over 2.2 million full-text articles, having grown by 13% over the past year. More than 1300 journals, including Nucleic Acids Research, deposit the full text of their articles in PMC, and 282 of these journals began depositing their data in the last year. Publisher participation in PMC requires a commitment to free access to full text, either immediately after publication or within a 12-month period. As a consequence of the mandatory NIH Public Access Policy that went into effect on April 7, 2008, PMC is also the repository for all final peer-reviewed manuscripts arising from research using NIH funds and submitted through the NIH Manuscript Submission System (NIHMS). All PMC articles are identified in PubMed search results and PMC itself can be searched using Entrez.
The NCBI bookshelf
Bookshelf, a part of the National Library of Medicine Literature Archive (www.ncbi.nlm.nih.gov/books/NBK51660/), offers users free access to the full text of more than 900 books, reports and databases in the life sciences and health care. In addition to new titles being added to Bookshelf each month (∼22 per month in 2011), updates are also made to existing titles to keep them current. Selected records in Bookshelf can be found in PubMed, under the Books and Documents label, and users can access the free full text in Bookshelf via the book icon in the PubMed record. Information in Bookshelf is linked to other NCBI resources, such as Gene, PubMed and PubChem, enabling the user to discover relevant information. In the past year, Bookshelf underwent a redesign to improve the overall display and information delivery of search results and book viewer pages. The redesign brings features to Bookshelf that PubMed users will find familiar, such as menus to control the display settings and downloads, consistent navigation in the search results pages and the Related Citations Discovery Component in the book viewer pages. Bookshelf also added a tool to enable easier browsing of titles by subject, resource type and publisher.
TAXONOMY
The NCBI taxonomy database is a central organizing principle for the Entrez biological databases and provides links to all data for each taxonomic node, from superkingdoms to subspecies. The taxonomy database reflects sequence data from almost 250 000 formally described species. This represents virtually all of the formally described species of prokaryotes, and 10% of the eukaryotes. The Taxonomy Browser (www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi) can be used to view the taxonomy tree or retrieve data from any of the Entrez databases for a particular organism or group.
DNA AND RNA
Reference sequences
The NCBI reference sequence (RefSeq) database (6) is a non-redundant set of curated and computationally derived sequences for transcripts, proteins and genomic regions. The number of nucleotide bases in the RefSeq collection has grown by 14% over the past year so that Release 48 (July, 2011) contains 164 billion bases representing over 12 200 organisms. RefSeq DNA and RNA sequences can be searched and retrieved from the Nucleotide database, and the complete RefSeq collection is available in the RefSeq directory on the NCBI FTP site.
Sequences from GenBank and other sources
Sequences from GenBank (1) can be searched in and retrieved from three Entrez databases: Nucleotide, Expressed Sequence Tag (EST) and Genome Survey Sequence (GSS) (specified as nuccore, nucest and nucgss within the E-utilities). The Nucleotide database contains all GenBank sequences except those within the EST or GSS GenBank divisions. The database also contains WGS sequences, Third Party Annotation (TPA) sequences and sequences imported from the Structure database. In addition, those sequences that have been submitted as part of a population, phylogenetic or environmental study are placed in the PopSet database.
The trace archive and SRA
The Trace Archive contains over 2 billion traces from gel and capillary electrophoresis sequencers. Data from more than 10 000 species are represented, including whole genomes of pathogens, organismal shotgun and BAC clone projects, and EST libraries. The Trace Archive was established after the conclusion of the Human Genome Sequencing Project, so only 12% of the traces are of human origin. The Trace Assembly Archive is a companion resource that contains placements of individual trace reads on a GenBank sequence. Using the sequence viewer, one can view multiple alignments of read placements at a given reference location. Many Influenza virus genomes are presented in this way.
The Sequence Read Archive (7) is a repository for sequencing data generated from next-generation sequencing technology and currently contains over 110 Terabasepairs (Tbps) of biological sequence data, including data mirrored from partner archives. Documentation on using and submitting data to the resource is available at www.ncbi.nlm.nih.gov/bookshelf/br.fcgi?book=helpsra. BLAST searches are offered for transcript and whole-genome sequence SRA data sets, and regular expression pattern matching against short reads of all types is possible. A version of the SRA has been deployed with dbGaP authorized access procedures to provide archive services for human sequencing data under privacy restrictions.
During 2011, NCBI encountered funding issues that resulted in a revised scope for SRA and Trace. NCBI will continue to handle raw sequencing data associated with RNA-Seq, ChIP-Seq and epigenomic data that are submitted to Gene expression omnibus (GEO); genomic and transcriptomic assemblies that are submitted to GenBank; and 16S ribosomal RNA data associated with metagenomics that are submitted to GenBank. In addition, many NIH Institutes are providing funding that allows for continued deposition of human genetic sequence data from their large studies into dbGaP and SRA. NCBI will continue to develop technologies for optimum storage and retrieval of raw sequencing data and their alignments to reference genomes.
PROTEINS
Reference sequences
In addition to genomic and transcript sequences, the RefSeq database (6) contains protein sequences that are curated and computationally derived from these DNA and RNA sequences. The number of amino acid residues in the RefSeq collection has grown by 23% over the past year so that Release 48 (July, 2011) contains 4.4 billion residues. RefSeq protein sequences can be searched and retrieved from the Protein database, and the complete RefSeq collection is available in the RefSeq directory on the NCBI FTP site.
Sequences from GenBank and other sources
As part of standard submission procedures, NCBI produces conceptual translations for any sequence in GenBank (1) that contains a coding sequence and places these protein sequences in the Protein database. In addition to these 29 million ‘GenPept’ sequences, the Protein database also contains sequences from TPA, UniProtKB/Swiss-Prot (8), the Protein Research Foundation (PRF) and the Protein Data Bank (PDB) (9).
Protein clusters
The Protein Clusters database contains over 620 000 sets of almost identical RefSeq proteins encoded by complete genomes from prokaryotes, eukaryotic organelles (mitochondria and chloroplasts), viruses and plasmids as well as from some protozoans and plants. The clusters are organized in a taxonomic hierarchy and are created based on reciprocal best-hit protein BLAST scores (10). These clusters are used as a basis for genome-wide comparison at NCBI as well as to provide simplified BLAST searches via Concise Microbial Protein BLAST (www.ncbi.nlm.nih.gov/genomes/prokhits.cgi). Protein clusters provide annotations, publications, domains, structures, external links and analysis tools, including multiple sequence alignments and phylogenetic trees.
HIV-1/human protein interaction database
The Division of Acquired Immunodeficiency Syndrome of the National Institute of Allergy and Infectious Diseases (NIAID), in collaboration with the Southern Research Institute and NCBI, maintains a comprehensive HIV Protein Interaction Database of documented interactions between HIV-1 proteins, host cell proteins, other HIV-1 proteins or proteins from disease organisms associated with HIV or AIDS (11). Summaries, including protein RefSeq accession numbers, Gene IDs, lists of interacting amino acids, brief descriptions of interactions, keywords and PubMed IDs for supporting journal articles are presented at www.ncbi.nlm.nih.gov/RefSeq/HIVInteractions/. All protein–protein interactions documented in the HIV Protein Interaction Database are listed in Gene reports in the HIV-1 protein interactions section.
BLAST SEQUENCE ANALYSIS
BLAST
The BLAST programs (12–14) perform sequence-similarity searches against a variety of nucleotide and protein databases, returning a set of gapped alignments with links to full sequence records as well as to related transcript clusters (UniGene), annotated gene loci (Gene), 3D structures (MMDB) or microarray studies (GEO). The NCBI web interface for BLAST allows users to assign titles to searches, to review recent search results and to save parameter sets in MyNCBI for future use. The basic BLAST programs are also available as standalone command line programs, as network clients, and as a local Web-server package at ftp.ncbi.nih.gov/blast/executables/LATEST/ (Table 2).
Table 2.
Selected NCBI software available for download
| Software | Available binaries | Category within this article |
|---|---|---|
| BLAST (stand alone) | Win, Mac, LINUX, Solaris | BLAST sequence analysis |
| BLAST (network client) | Win, Mac, LINUX, Solaris | BLAST sequence analysis |
| BLAST (web server) | Mac, LINUX, Solaris | BLAST sequence analysis |
| CD-Tree | Win, Mac | Domains and structures |
| Cn3D | Win, Mac | Domains and structures |
| PC3D | Win, Mac, LINUX | Chemicals and bioassays |
| gene2xml | Win, Mac, LINUX, Solaris | Genes and expression |
| Genome Workbench | Win, Mac, LINUX | Genomes |
| splign | LINUX, Solaris | Genomes |
| tbl2asn | Win, Mac, LINUX, Solaris | Genomes |
BLAST databases
The default database for nucleotide BLAST searches (Human genomic plus transcript) contains human RefSeq transcript and genomic sequences arising from the NCBI annotation of the human genome. Searches of this database generate a tabular display that partitions the BLAST hits by sequence type (genomic or transcript) and allows sorting by BLAST score, percent identity within the alignment and the percent of the query sequence contained in the alignment. A similar database is available for mouse. Several other databases are also available and are described in links from the BLAST input form. Each of these databases can be limited to an arbitrary taxonomic node or those records satisfying any Entrez query.
For proteins, the default database (nr) is a non-redundant set of all CDS translations from GenBank along with all RefSeq, UniProtKB/Swiss-Prot, PDB and PRF proteins. Subsets of this database are also available, such as the PDB or UniProtKB/Swiss-Prot sequences, along with separate databases for sequences from patents and environmental samples. Like the nucleotide databases, these collections can be limited by taxonomy or an arbitrary Entrez query.
BLAST output formats
Standard BLAST output formats include the default pairwise alignment, several query-anchored multiple sequence alignment formats, an easily-parsable Hit Table and a report that organizes the BLAST hits by taxonomy. A ‘pairwise with identities’ mode better highlights differences between the query and a target sequence. A tree view option for the Web BLAST service creates a dendrogram that clusters sequences according to their distances from the query sequence. Each alignment returned by BLAST is scored and assigned a measure of statistical significance, called the Expectation Value (E-value). The alignments returned can be limited by an E-value threshold or range.
Genomic BLAST
NCBI maintains Genomic BLAST services for more than 120 organisms shown in the Map Viewer. By default, genomic BLAST searches the genomic sequence of an organism, but additional databases are also available, such as the nucleotide and protein RefSeqs annotated on the genomic sequence, as well as sets of sequences such as ESTs that are mapped to the genomic sequence. The default search program for the NCBI Genomic BLAST pages is MegaBLAST (15), a faster version of standard nucleotide BLAST designed to find alignments between nearly identical sequences, typically from the same species. For rapid cross-species nucleotide queries, NCBI offers Dis-contiguous MegaBLAST, which uses a non-contiguous word match (16) as the nucleus for its alignments. Dis-contiguous MegaBLAST is far more rapid than a translated search such as blastx, yet maintains a competitive degree of sensitivity when comparing coding regions.
Primer-BLAST
Primer-BLAST is a tool for designing and analyzing PCR primers based on the existing program Primer3 (17) that designs PCR primers given a template DNA sequence. Primer-BLAST extends this functionality by running a BLAST search against a chosen database with the designed primers as queries, and then returns only those primer pairs specific to the desired target, in that they do not generate valid PCR products on unintended targets. Users can also specify a forward or reverse primer in addition to a DNA template, in which case the other primer will be designed and analyzed. If both primers are specified along with a template, the tool performs only the final BLAST analysis. Users may also enter two primers without a template, in which case the BLAST analysis will display those templates in the chosen database that best match the primer pair. The available databases range from RefSeq mRNA or genomic sets for 1 of 12 model organisms to the entire BLAST nr database. A new optional graphic result display is now available for viewing more details about the primers.
COBALT
COBALT (18) is a multiple alignment algorithm that finds a collection of pairwise constraints derived from both the NCBI Conserved Domain database (CDD) and the sequence similarity programs RPS-BLAST, BLASTP and PHI-BLAST. These pair-wise constraints are then incorporated into a progressive multiple alignment. COBALT searches can be launched either from a BLASTP result page or from the main COBALT search page, where either FASTA sequences or accessions (or a combination thereof) may be entered into the query sequence box. Links at the top of the COBALT report provide access to a phylogenetic tree view of the multiple alignments and allow users either to launch a modified search or download the alignment in several popular formats.
GENES AND EXPRESSION
Gene
Gene (19) provides an interface to curated sequences and descriptive information about genes with links to NCBI's Map Viewer, Evidence Viewer, Model Maker, BLink, protein domains from the CDD, and other gene-related resources. Gene contains data for almost 8 million genes from over 8400 organisms. These data are accumulated and maintained through several international collaborations in addition to curation by in-house staff. Links within Gene to the newest citations in PubMed are maintained by curators and provided as Gene References into Function (GeneRIF). The complete Gene data set, as well as organism-specific subsets, is available in the compact NCBI Abstract Syntax Notation One (ASN.1) format on the NCBI FTP site. The gene2xml tool converts the native Gene ASN.1 format into XML and is available at ftp.ncbi.nih.gov/toolbox/ncbi_tools/converters/by_program/gene2xml/.
RefSeqGene
In collaboration with Locus Reference Genomic (www.lrg-sequence.org), RefSeqGene provides stable, standard human genomic sequences annotated with standard mRNAs for well-characterized human genes (6). RefSeqGene records are part of the RefSeq collection and are created in consultation with authoritative locus-specific databases or other experts on particular loci and provide a stable genomic sequence for establishing numbering systems for exons and introns and for reporting and identifying genomic variants, especially those of clinical importance (20). By default, a RefSeqGene record begins 5 kb upstream of the first exon of the gene and ends 2 kb downstream of the final exon, but those positions will be adjusted on request. A RefSeqGene sequence may differ from the current genomic build so as to reflect standard alleles. RefSeqGene records can be retrieved from Nucleotide using the query ‘refseqgene[keyword]’, are available on corresponding Gene reports and can be downloaded from ftp.ncbi.nih.gov/refseq/H_sapiens/RefSeqGene.
The conserved CDS database (CCDS)
The CCDS project is a collaborative effort among NCBI, the European Bioinformatics Institute, the Wellcome Trust Sanger Institute (WTSI) and University of California, Santa Cruz (UCSC) to identify a set of human and mouse protein coding regions that are consistently annotated and of high quality (21). The collaborators prepare the CCDS set by comparing the annotations they have independently determined and then identifying those coding regions that have identical coordinates on the genome. Those regions that pass quality evaluations are then added to the CCDS set. To date, the CCDS database contains over 25 500 human and 22 100 mouse CDS annotations. The web interface to the CCDS allows searches by gene or sequence identifiers and provides links to Gene, record revision histories, transcript and protein sequences as well as gene views in Map Viewer, the Ensemble Genome Browser, the UCSC Genome Browser and the Sanger Institute Vega Browser. The CCDS sequence data are available at ftp.ncbi.nlm.nih.gov/pub/CCDS/.
GEO
GEO (22) is a data repository and retrieval system for high-throughput functional genomic data generated by microarray and next-generation sequencing technologies. In addition to gene expression data, GEO accepts other categories of experiments including studies of genome copy number variation, genome–protein interaction surveys and methylation profiling studies. The repository can capture fully annotated raw and processed data, enabling compliance with major community-derived scientific reporting standards such as ‘Minimum Information About a Microarray Experiment’ (MIAME) (23,24). Several data deposit options and formats are supported, including web forms, spreadsheets, XML and plain text. GEO data are housed in two Entrez databases: GEO Profiles, which contains quantitative gene expression measurements for one gene across an experiment, and GEO Data sets, which contains entire experiments. Currently, the GEO database hosts over 25 000 studies submitted by 10 000 laboratories and comprising 625 000 samples and 50 billion individual abundance measurements for over 1600 organisms. The distribution of study types contained within GEO can be viewed at www.ncbi.nlm.nih.gov/geo/summary/.
UniGene and ProtEST
UniGene (25) is a system for partitioning transcript sequences (including ESTs) from GenBank into a non-redundant set of clusters, each of which contains sequences that seem to be produced by the same transcription locus. UniGene clusters are created for all organisms for which there are 70 000 or more ESTs in GenBank, and currently the database includes clusters for 139 eukaryotes. UniGene databases are updated weekly with new EST sequences, and bimonthly with newly characterized sequences. As an aid to identifying a UniGene cluster, ProtEST presents precomputed BLAST alignments between protein sequences from model organisms and the six-frame translations of nucleotide sequences in UniGene.
HomoloGene
HomoloGene is a system that automatically detects homologs, including paralogs and orthologs, among the genes of 20 completely sequenced eukaryotic genomes. HomoloGene reports include homology and phenotype information drawn from Online Mendelian Inheritance in Man (OMIM) (26), Mouse Genome Informatics (MGI) (27), Zebrafish Information Network (ZFIN) (28), Saccharomyces Genome Database (SGD) (29), Clusters of Orthologous Groups (COG) (30) and FlyBase (31). Information about the HomoloGene build procedure is provided at www.ncbi.nlm.nih.gov/HomoloGene/HTML/homologene_buildproc.html. The HomoloGene Downloader, appearing under the ‘Download’ link in HomoloGene displays, retrieves transcript, protein or genomic sequences for the genes in a HomoloGene group; in the case of genomic sequence, upstream and downstream regions may be specified.
Probe
The Probe database is a public registry of nucleic acid reagents designed for use in a wide variety of biomedical research applications, together with information on reagent distributors, probe effectiveness and computed sequence similarities. The Probe database archives 10.4 million probe sequences, among them probes for genotyping, SNP discovery, gene expression, gene silencing and gene mapping. The probe database also provides submission templates to simplify the process of depositing data (www.ncbi.nlm.nih.gov/genome/probe/doc/Submitting.shtml).
Biosystems
The Biosystems database collects together molecules that interact in a biological system, such as a biochemical pathway or disease. Currently Biosystems receives data from the Kyoto Encyclopedia of Genes and Genomes (KEGG) (32–34), BioCyc (35), Reactome (36), the Pathway Interaction Database (37) and WikiPathways (38,39). These source databases provide diagrams of pathways that display the various components with their substrates and products, as well as links to relevant literature. In addition to being linked to such literature in PubMed, each component within a Biosystem record is also linked to the corresponding records in Gene and Protein, while the substrates and products are linked to records in PubChem (see below) so that the Biosystem record centralizes NCBI data related to the pathway, greatly facilitating computation on such systems.
GENOMES
Genome
The Genome database (40) provides access to genomic sequences from the RefSeq collection and is a convenient portal both for retrieving such sequences from multiple organisms and for viewing small genomes, such as those from prokaryotes. Currently the database contains complete genomes for more than 1700 microbes and 2600 viruses, as well as for over 2800 eukaryotic organelles. For higher eukaryotes, the Genome database includes complete genomes for 37 species, as well as data from over 1100 other genome-sequencing projects. More than 11% of the 13 800 total sequences were added in the past year. For higher eukaryotes, Genome provides direct links to the NCBI Map Viewer; for prokaryotes, viruses and eukaryotic organelles, specialized viewers and BLAST pages are available. The Plant Genomes Central Web page serves as a portal to completed plant genomes, to information on plant genome-sequencing projects or to other resources at NCBI such as the plant Genomic BLAST pages or Map Viewer.
The NCBI Map Viewer displays genome assemblies, genetic and physical markers and the results of annotation and other analyses using sets of aligned maps for 121 organisms. The available maps vary by organism and may include cytogenetic maps, physical maps and a variety of sequence-based maps. Maps from multiple organisms or multiple assemblies for the same organism can be displayed in a single view. Map Viewer also can display to previous genome builds and can produce convenient formats for downloading data.
NCBI's Genome Workbench is a stand-alone application (Table 2) for sequence and genomic evaluation, offering tools for visualization and analysis, including integrated graphical views of sequences and alignments, text and tabular displays of annotation, and common sequence analysis tools, including BLAST, MUSCLE, and Splign. Genome Workbench offers the power of computation on a user's own computer, and can easily combine private data with data available for public retrieval. The most recent version, 2.4.0, contains several new features and bug fixes outlined at www.ncbi.nlm.nih.gov/projects/gbench/release-notes.html. In addition, video tutorials are available on the NCBI YouTube channel (www.youtube.com/NCBINLM).
The Genome Reference Consortium
The Genome Reference Consortium (GRC) (www.ncbi.nlm.nih.gov/projects/genome/assembly/grc/) is an international collaboration between WTSI, the Genome Institute at Washington University, EMBL and NCBI that aims to produce assemblies of higher eukaryotic genomes that best reflect complex allelic diversity consistent with currently available data. The GRC has produced a major update for human (GRCh37) (PMID 19468303) and is currently curating the reference assemblies of mouse (MGSCv37) and Zebrafish (Zv9, www.sanger.ac.uk/Projects/D_rerio/Zv9_assembly_information.shtml); a major update of the mouse assembly will be available from the GRC in December 2011 (GRCm38). Between major assembly releases, the GRC provides minor ‘patch’ releases that provide additional sequence scaffolds that either correct errors in the assembly (fix patches) or add an alternate loci (novel patches). In the next major assembly release, the changes represented by the fix patches will be incorporated into the new assembly, and the fix patches themselves will be removed from the release. Novel patches will become alternate loci integrated into the new assembly. The NCBI Map Viewer provides views of the most recent GRC releases for human and mouse and for Zv9 for zebrafish, and the GRC data are available for download from the NCBI FTP site at links provided on the GRC web pages.
Epigenomics
The Epigenomics database collects data from studies examining epigenetic features such as posttranslational modifications of histone proteins, genomic DNA methylation, chromatin organization and the expression of non-coding regulatory RNA (41). Raw data from these experiments, together with extensive meta-data, are stored in the GEO and SRA databases. The Epigenomics resource provides a higher level view, allowing users to search and browse the data based on biological attributes such as cell type, tissue type, differentiation stage and heath status, among many others. Data have been premapped to genomic coordinates (to make ‘genome tracks’), so users are not required to be familiar with or manipulate the raw data. Tracks may be visualized in either the NCBI or UCSC genome viewers or may be downloaded to the user's computer for local analysis. Data from the Roadmap Epigenomics project, which are currently being hosted at GEO (www.ncbi.nlm.nih.gov/geo/roadmap/epigenomics/), are being mirrored and are available for viewing and downloading.
Database of genomic structural variation
The Database of Genomic Structural Variation (dbVar) is an archive of large-scale genomic variants such as insertions, deletions, translocations and inversions. Currently dbVar (42) contains over 60 studies from human, rhesus macaque, chimpanzee, mouse, dog, fruit fly and pig, and accepts data derived from several methods including computational sequence analysis and microarray experiments. Each of the over 800 000 variants is linked to a graphical view showing its genomic context. Symbols for genes within variant regions are now displayed on search results, and users can also search for such genes directly in dbVar.
Influenza genome resources
The Influenza Genome Sequencing Project (IGSP) (43) provides researchers with a growing collection of over 60 000 virus sequences essential to the identification of the genetic determinants of influenza pathogenicity. NCBI's Influenza Virus Resource links the IGSP project data via PubMed to the most recent scientific literature on influenza as well as to a number of online analysis tools and databases. These databases include NCBI's Influenza Virus Sequence Database, comprised of over 190 000 influenza sequences in the GenBank and RefSeq databases, as well as other Entrez databases containing 210 000 influenza protein sequences, 200 influenza protein structures and 700 influenza population studies. An online influenza genome annotation tool analyzes a novel sequence and produces output in a ‘feature table’ format that can be used by NCBI's GenBank submission tools such as tbl2asn (1).
NCBI now also provides the Virus Variation resource (http://www.ncbi.nlm.nih.gov/genomes/VirusVariation/genomes/VirusVariation/) that extends services available for Influenza to other viruses, such as the Dengue virus. Virus variation provides a portal for retrieving, downloading, analyzing and annotating virus sequences using pages customized to unique aspects of viral sequence data, including genotype, severity of the resulting disease and the year a sample was collected.
GENETICS AND MEDICINE
The Database of Genotypes and Phenotypes (dbGaP)
The Database of Genotypes and Phenotypes (dbGaP) (44) archives, distributes and supports submission of data that correlate genomic characteristics with observable traits. This database is a designated NIH repository for NIH-funded genome-wide association study (GWAS) results (grants.nih.gov/grants/gwas/index.htm). The dbGaP collection contains over 150 studies, each of which can be browsed by name or disease.
To protect the confidentiality of study subjects, dbGaP accepts only deidentified data and requires investigators to go through an authorization process to access individual-level data. Study documents, protocols and subject questionnaires are available without restriction. Authorized access data distributed to primary investigators for use in approved research projects includes deidentified phenotypes and genotypes for individual study subjects, pedigrees and some precomputed associations between genotype and phenotype.
dbSNP
The Database of Short Genetic Variations (dbSNP) (45) is a repository for a variety of short genetic variations such as single nucleotide polymorphisms (SNPs), insertions/deletions, microsatellites and non-polymorphic variants. dbSNP also stores common and rare variations along with their genotypes and allele frequencies, and includes clinically significant human variations as well as benign polymorphisms. In addition to archiving the variant and sequence position, dbSNP maintains information about population-specific allele frequencies and individual genotypes, validation status, availability of significant genome-wide association results and PubMed citations for clustered reference records (rs#). dbSNP also aggregates user assertions for these reference records, including its clinical significance, the likelihood that the variation is a false positive and whether the allele origin is germline or somatic. These aggregated assertions may arise from multiple submitters with different levels of experimental support and may, therefore, conflict. dbSNP does not independently verify assertions and cannot endorse their accuracy.
dbSNP integrates information about genetic variants with clinical relevance in collaboration with locus-specific databases (LSDBs) and clinical diagnostic laboratories. An automated web submission portal is available to facilitate the submission of LSDB/clinical variant information and to support variant descriptions using the HGVS standards applied to a RefSeq standard sequence. Users can search and annotate existing variations or submit novel ones, either as a single variation (www.ncbi.nlm.nih.gov/projects/SNP/tranSNP/tranSNP.cgi) or as a batch (www.ncbi.nlm.nih.gov/projects/SNP/tranSNP/VarBatchSub.cgi).
GeneReviews and GeneTests
NCBI hosts GeneReviews and GeneTests, two resources developed by a team led by Roberta A. Pagon, MD at the University of Washington. GeneReviews (www.ncbi.nlm.nih.gov/bookshelf/br.fcgi?book=gene) is a compendium of continually updated, expert-authored and peer-reviewed disease descriptions that relate genetic testing to the diagnosis, management and genetic counseling of patients and families with specific inherited conditions (46,47). These reviews can be searched via the GeneReviews tab at the GeneTests home page (www.ncbi.nlm.nih.gov/sites/GeneTests/), NCBI's Bookshelf site, NCBI's All Databases interface or major web search engines.
The GeneTests laboratory directory and clinic directory list information voluntarily provided by laboratories about their tests and by genetics clinics about their clinical genetics services. As appropriate, users can search by a disease name, gene symbol, protein name, clinical genetics service and information about a lab/clinic, such as its name, director and location. Clinics in the USA can also be found via a map-based search. Together, GeneReviews and the GeneTests directories support the integration of information on genetic disorders and genetic testing into a single resource to facilitate the care of patients and families with inherited conditions.
Online Mendelian Inheritance in Animals
OMIA is a database of genes, inherited disorders and traits in animal species other than human and mouse, and is authored by Professor Frank Nicholas and colleagues (48) of the University of Sydney, Australia. The database holds 2800 records containing textual information and references, as well as links to relevant records from OMIM, PubMed and Gene.
Database cluster for routine clinical applications: dbMHC, dbLRC, dbRBC
dbMHC focuses on the Major Histocompatibility Complex (MHC) and contains sequences and frequency distributions for alleles of the MHC, an array of genes that play a central role in the success of organ transplants and an individual's susceptibility to infectious diseases. dbMHC also contains HLA genotype and clinical outcome information on hematopoietic cell transplants performed worldwide. dbLRC offers a comprehensive collection of alleles of the leukocyte receptor complex with an emphasis on KIR genes. dbRBC represents data on genes and their sequences for red blood cell antigens or blood groups. It hosts the blood group antigen gene mutation database (49) and integrates it with resources at NCBI. dbRBC provides general information on individual genes and access to the ISBT allele nomenclature of blood group alleles. All three databases dbMHC, dbLRC and dbRBC provide multiple sequence alignments, analysis tools to interpret homozygous or heterozygous sequencing results (50) and tools for DNA probe alignments.
CHEMICALS AND BIOASSAYS
PubChem (60) is the informatics backbone for the NIH Roadmap Initiative on molecular libraries and focuses on the chemical, structural and biological properties of small molecules, in particular their roles as diagnostic and therapeutic agents. A suite of three Entrez databases, PCSubstance, PCCompound and PCBioAssay, contain the structural and bioactivity data of the PubChem project. The databases include records for 85 million substances containing 30 million unique chemical structures, and 2.1 million of these substances have bioactivity data in at least one of the 504 000 PubChem BioAssays. PubChem also provides a diverse set of three-dimensional (3D) conformers for 90% of the records in the PubChem compound database. A viewing application, PC3D, is available to view both individual conformers and overlays of similar conformers. The PubChem databases link not only to other Entrez databases such as PubMed and PMC but also to structure and protein to provide a bridge between the macromolecules of genomics and the small organic molecules of cellular metabolism. The PubChem databases are searchable using text queries as well as structural queries based on chemical SMILES, formulas or chemical structures provided in a variety of formats. The PubChem Sketcher, an online structure-drawing tool provides a simple way to construct a structure-based search (pubchem.ncbi.nlm.nih.gov/search/search.cgi).
DOMAINS AND STRUCTURES
The Molecular Modeling Database
The NCBI Molecular Modeling Database (MMDB) (51) contains experimentally determined coordinate sets from the Protein Data Bank (9), augmented with domain annotations and links to relevant literature, protein and nucleotide sequences, chemicals (PDB heterogens) and conserved domains in CDD (52). The structure summary pages for individual MMDB records were recently redesigned (see above) and display these links along with thumbnail images of structures that link to interactive views of the data in Cn3D (53), the NCBI structure and alignment viewer. Compact structural domains within protein structures are annotated on protein chains, and these graphic annotations link to structural neighbors computed by the VAST algorithm (54,55). Users can access MMDB structures either through direct text searches or through the ‘Related Structures’ link provided for all protein records.
CDD and CDART
The CDD (56) contains over 40 000 PSI-BLAST-derived position-specific score matrices representing domains taken from the simple modular architecture research tool (Smart) (57), Pfam (58), TIGRFAM (59), and from domain alignments derived from COGs and Protein Clusters. In addition, CDD includes 3300 superfamily records, each of which contains a set of CDs from one or more source databases that generate overlapping annotation on the same protein sequences. The NCBI Conserved Domain Search (CD-Search) service locates conserved domains within a protein sequence, and these results are available for all records in the Protein database through the ‘Identify Conserved Domains’ link in the upper right of a sequence record. Wherever possible, protein sequences with known 3D structures are included in CD alignments, which can be viewed along with these structures and also edited within Cn3D. CD alignments can be viewed online, edited or created de novo using CDTree (Table 2). CDTree uses PSI-BLAST to add new sequences to an existing CD alignment and provides an interface for exploring phylogenetic trends in domain architecture and for building hierarchies of alignment-based protein domains. The Conserved Domain Architecture Retrieval Tool (CDART) searches protein databases with a query sequence and returns the domain architectures of database proteins containing the query domain.
FOR FURTHER INFORMATION
The resources described here include documentation, other explanatory material and references to collaborators and data sources on the respective websites. An alphabetical list of NCBI resources is available from a link in the upper left of the NCBI home page. The NCBI Help Manual and the NCBI Handbook, both available as links in the common page footer, describe the principal NCBI resources in detail. The NCBI Education page (www.ncbi.nlm.nih.gov/Education/) lists links to documentation, tutorials and educational tools along with links to outreach initiatives including Discovery Workshops, webinars and upcoming conference exhibits. The Education page, along with the standard NCBI page footer, contains links to the NCBI pages on Facebook, Twitter and YouTube. Several new training videos produced in the past year have been added to YouTube. A user-support staff is available to answer questions at info@ncbi.nlm.nih.gov. Updates on NCBI resources and database enhancements are described in the NCBI News newsletter (http://www.ncbi.nlm.nih.gov/books/NBK1969/). In addition, NCBI offers several mailing lists that provide updates on services and databases (www.ncbi.nlm.nih.gov/Sitemap/Summary/email_lists.html), as well as RSS feeds (www.ncbi.nlm.nih.gov/feed/).
FUNDING
Funding for open access charge: Intramural Research Program of the National Institutes of Health, National Library of Medicine.
Conflict of interest statement. None declared.
REFERENCES
- 1.Benson DA, Karsch-Mizrachi I, Clark K, Lipman DJ, Ostell J, Sayers EW. GenBank. Nucleic Acids Res. 2012;40:D48–D53. doi: 10.1093/nar/gkr1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barrett T, Clark K, Gevorgyan R, Gorelenkov V, Gribov E, Kimelman M, Mizrachi I, Pruitt KD, Resenchuk S, Tatusova T, et al. BioProject and BioSample databases at NCBI: facilitating capture and organization of metadata. Nucleic Acids Res. 2012;40:D57–D63. doi: 10.1093/nar/gkr1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schuler GD, Epstein JA, Ohkawa H, Kans JA. Entrez: molecular biology database and retrieval system. Methods Enzymol. 1996;266:141–162. doi: 10.1016/s0076-6879(96)66012-1. [DOI] [PubMed] [Google Scholar]
- 4.Sewell W. Medical subject headings in medlars. Bull. Med. Libr. Assoc. 1964;52:164–170. [PMC free article] [PubMed] [Google Scholar]
- 5.Sequeira E. PubMed central—3 years old and growing stronger. ARL. 2003;228:5–9. [Google Scholar]
- 6.Pruitt KD, Tatusova T, Klimke W, Maglott DR. NCBI reference sequences: current status, policy and new initiatives. Nucleic Acids Res. 2009;37:D32–D36. doi: 10.1093/nar/gkn721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shumway M, Cochrane G, Sugawara H. Archiving next generation sequencing data. Nucleic Acids Res. 2010;38:D870–D871. doi: 10.1093/nar/gkp1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Magrane M, Consortium U. UniProt knowledgebase: a hub of integrated protein data. Database. 2011;2011:bar009. doi: 10.1093/database/bar009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Berman H, Henrick K, Nakamura H, Markley JL. The worldwide Protein Data Bank (wwPDB): ensuring a single, uniform archive of PDB data. Nucleic Acids Res. 2007;35:D301–D303. doi: 10.1093/nar/gkl971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Klimke W, Agarwala R, Badretdin A, Chetvernin S, Ciufo S, Fedorov B, Kiryutin B, O'Neill K, Resch W, Resenchuk S, et al. The national center for biotechnology information's protein clusters database. Nucleic Acids Res. 2009;37:D216–D223. doi: 10.1093/nar/gkn734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fu W, Sanders-Beer BE, Katz KS, Maglott DR, Pruitt KD, Ptak RG. Human immunodeficiency virus type 1, human protein interaction database at NCBI. Nucleic Acids Res. 2009;37:D417–D422. doi: 10.1093/nar/gkn708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 13.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ye J, McGinnis S, Madden TL. BLAST: improvements for better sequence analysis. Nucleic Acids Res. 2006;34:W6–W9. doi: 10.1093/nar/gkl164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang Z, Schwartz S, Wagner L, Miller W. A greedy algorithm for aligning DNA sequences. J. Comput. Biol. 2000;7:203–214. doi: 10.1089/10665270050081478. [DOI] [PubMed] [Google Scholar]
- 16.Ma B, Tromp J, Li M. PatternHunter: faster and more sensitive homology search. Bioinformatics. 2002;18:440–445. doi: 10.1093/bioinformatics/18.3.440. [DOI] [PubMed] [Google Scholar]
- 17.Rozen S, Skalestsky HJ. Primer3 on the WWW for General Users and for Biologist Programmers. In: Krawetz S, Misener S, editors. Bioinformatics Methods and Protocols: Methods in Molecular Biology. Totowa, NJ: Humana Press; 2000. pp. 365–386. [DOI] [PubMed] [Google Scholar]
- 18.Papadopoulos JS, Agarwala R. COBALT: constraint-based alignment tool for multiple protein sequences. Bioinformatics. 2007;23:1073–1079. doi: 10.1093/bioinformatics/btm076. [DOI] [PubMed] [Google Scholar]
- 19.Maglott D, Ostell J, Pruitt KD, Tatusova T. Entrez Gene: gene-centered information at NCBI. Nucleic Acids Res. 2011;39:D52–D57. doi: 10.1093/nar/gkq1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gulley ML, Braziel RM, Halling KC, Hsi ED, Kant JA, Nikiforova MN, Nowak JA, Ogino S, Oliveira A, Polesky HF, et al. Clinical laboratory reports in molecular pathology. Arch. Pathol. Lab. Med. 2007;131:852–863. doi: 10.5858/2007-131-852-CLRIMP. [DOI] [PubMed] [Google Scholar]
- 21.Pruitt KD, Harrow J, Harte RA, Wallin C, Diekhans M, Maglott DR, Searle S, Farrell CM, Loveland JE, Ruef BJ, et al. The consensus coding sequence (CCDS) project: identifying a common protein-coding gene set for the human and mouse genomes. Genome Res. 2009;19:1316–1323. doi: 10.1101/gr.080531.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barrett T, Troup DB, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH, Sherman PM, et al. NCBI GEO: archive for functional genomics data sets—10 years on. Nucleic Acids Res. 2011;39:D1005–D1010. doi: 10.1093/nar/gkq1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brazma A, Hingamp P, Quackenbush J, Sherlock G, Spellman P, Stoeckert C, Aach J, Ansorge W, Ball CA, Causton HC, et al. Minimum information about a microarray experiment (MIAME)—toward standards for microarray data. Nat. Genet. 2001;29:365–371. doi: 10.1038/ng1201-365. [DOI] [PubMed] [Google Scholar]
- 24.Whetzel PL, Parkinson H, Causton HC, Fan L, Fostel J, Fragoso G, Game L, Heiskanen M, Morrison N, Rocca-Serra P, et al. The MGED ontology: a resource for semantics-based description of microarray experiments. Bioinformatics. 2006;22:866–873. doi: 10.1093/bioinformatics/btl005. [DOI] [PubMed] [Google Scholar]
- 25.Schuler GD. Pieces of the puzzle: expressed sequence tags and the catalog of human genes. J. Mol. Med. 1997;75:694–698. doi: 10.1007/s001090050155. [DOI] [PubMed] [Google Scholar]
- 26.Amberger J, Bocchini CA, Scott AF, Hamosh A. McKusick's online Mendelian inheritance in man (OMIM) Nucleic Acids Res. 2009;37:D793–D796. doi: 10.1093/nar/gkn665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eppig JT, Blake JA, Bult CJ, Kadin JA, Richardson JE. The mouse genome database (MGD): new features facilitating a model system. Nucleic Acids Res. 2007;35:D630–D637. doi: 10.1093/nar/gkl940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sprague J, Bayraktaroglu L, Clements D, Conlin T, Fashena D, Frazer K, Haendel M, Howe DG, Mani P, Ramachandran S, et al. The zebrafish information network: the zebrafish model organism database. Nucleic Acids Res. 2006;34:D581–D585. doi: 10.1093/nar/gkj086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hong EL, Balakrishnan R, Dong Q, Christie KR, Park J, Binkley G, Costanzo MC, Dwight SS, Engel SR, Fisk DG, et al. Gene ontology annotations at SGD: new data sources and annotation methods. Nucleic Acids Res. 2008;36:D577–D581. doi: 10.1093/nar/gkm909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tatusov RL, Fedorova ND, Jackson JD, Jacobs AR, Kiryutin B, Koonin EV, Krylov DM, Mazumder R, Mekhedov SL, Nikolskaya AN, et al. The COG database: an updated version includes eukaryotes. BMC Bioinformatics. 2003;4:41. doi: 10.1186/1471-2105-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Crosby MA, Goodman JL, Strelets VB, Zhang P, Gelbart WM. FlyBase: genomes by the dozen. Nucleic Acids Res. 2007;35:D486–D491. doi: 10.1093/nar/gkl827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kanehisa M, Araki M, Goto S, Hattori M, Hirakawa M, Itoh M, Katayama T, Kawashima S, Okuda S, Tokimatsu T, et al. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008;36:D480–D484. doi: 10.1093/nar/gkm882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kanehisa M, Goto S, Hattori M, Aoki-Kinoshita KF, Itoh M, Kawashima S, Katayama T, Araki M, Hirakawa M. From genomics to chemical genomics: new developments in KEGG. Nucleic Acids Res. 2006;34:D354–D357. doi: 10.1093/nar/gkj102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Keseler IM, Bonavides-Martinez C, Collado-Vides J, Gama-Castro S, Gunsalus RP, Johnson DA, Krummenacker M, Nolan LM, Paley S, Paulsen IT, et al. EcoCyc: a comprehensive view of Escherichia coli biology. Nucleic Acids Res. 2009;37:D464–D470. doi: 10.1093/nar/gkn751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Matthews L, Gopinath G, Gillespie M, Caudy M, Croft D, de Bono B, Garapati P, Hemish J, Hermjakob H, Jassal B, et al. Reactome knowledgebase of human biological pathways and processes. Nucleic Acids Res. 2009;37:D619–D622. doi: 10.1093/nar/gkn863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schaefer CF, Anthony K, Krupa S, Buchoff J, Day M, Hannay T, Buetow KH. PID: the pathway interaction database. Nucleic Acids Res. 2009;37:D674–D679. doi: 10.1093/nar/gkn653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kelder T, Pico AR, Hanspers K, van Iersel MP, Evelo C, Conklin BR. Mining biological pathways using WikiPathways web services. PLoS One. 2009;4:e6447. doi: 10.1371/journal.pone.0006447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pico AR, Kelder T, van Iersel MP, Hanspers K, Conklin BR, Evelo C. WikiPathways: pathway editing for the people. PLoS Biol. 2008;6:e184. doi: 10.1371/journal.pbio.0060184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tatusova TA, Karsch-Mizrachi I, Ostell JA. Complete genomes in WWW Entrez: data representation and analysis. Bioinformatics. 1999;15:536–543. doi: 10.1093/bioinformatics/15.7.536. [DOI] [PubMed] [Google Scholar]
- 41.Fingerman IM, McDaniel L, Zhang X, Ratzat W, Hassan T, Jiang Z, Cohen RF, Schuler GD. NCBI Epigenomics: a new public resource for exploring epigenomic data sets. Nucleic Acids Res. 2011;39:D908–D912. doi: 10.1093/nar/gkq1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Church DM, Lappalainen I, Sneddon TP, Hinton J, Maguire M, Lopez J, Garner J, Paschall J, Dicuccio M, Yaschenko E, et al. Public data archives for genomic structural variation. Nat Genet. 2010;42:813–814. doi: 10.1038/ng1010-813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ghedin E, Sengamalay NA, Shumway M, Zaborsky J, Feldblyum T, Subbu V, Spiro DJ, Sitz J, Koo H, Bolotov P, et al. Large-scale sequencing of human influenza reveals the dynamic nature of viral genome evolution. Nature. 2005;437:1162–1166. doi: 10.1038/nature04239. [DOI] [PubMed] [Google Scholar]
- 44.Manolio TA, Rodriguez LL, Brooks L, Abecasis G, Ballinger D, Daly M, Donnelly P, Faraone SV, Frazer K, Gabriel S, et al. New models of collaboration in genome-wide association studies: the genetic association information network. Nat. Genet. 2007;39:1045–1051. doi: 10.1038/ng2127. [DOI] [PubMed] [Google Scholar]
- 45.Sherry ST, Ward MH, Kholodov M, Baker J, Phan L, Smigielski EM, Sirotkin K. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pagon RA. GeneTests: an online genetic information resource for health care providers. J. Med. Libr. Assoc. 2006;94:343–348. [PMC free article] [PubMed] [Google Scholar]
- 47.Waggoner DJ, Pagon RA. Internet resources in medical genetics. Curr. Protoc. Hum. Genet. 2009 doi: 10.1002/0471142905.hg0912s62. Chapter 9, Unit 9 12. [DOI] [PubMed] [Google Scholar]
- 48.Lenffer J, Nicholas FW, Castle K, Rao A, Gregory S, Poidinger M, Mailman MD, Ranganathan S. OMIA (Online Mendelian Inheritance in Animals): an enhanced platform and integration into the Entrez search interface at NCBI. Nucleic Acids Res. 2006;34:D599–D601. doi: 10.1093/nar/gkj152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Blumenfeld OO, Patnaik SK. Allelic genes of blood group antigens: a source of human mutations and cSNPs documented in the blood group antigen gene mutation database. Hum. Mutat. 2004;23:8–16. doi: 10.1002/humu.10296. [DOI] [PubMed] [Google Scholar]
- 50.Helmberg W, Dunivin R, Feolo M. The sequencing-based typing tool of dbMHC: typing highly polymorphic gene sequences. Nucleic Acids Res. 2004;32:W173–W175. doi: 10.1093/nar/gkh424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang Y, Addess KJ, Chen J, Geer LY, He J, He S, Lu S, Madej T, Marchler-Bauer A, Thiessen PA, et al. MMDB: annotating protein sequences with Entrez's 3D-structure database. Nucleic Acids Res. 2007;35:D298–D300. doi: 10.1093/nar/gkl952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Marchler-Bauer A, Lu S, Anderson JB, Chitsaz F, Derbyshire MK, DeWeese-Scott C, Fong JH, Geer LY, Geer RC, Gonzales NR, et al. CDD: a conserved domain database for the functional annotation of proteins. Nucleic Acids Res. 2011;39:D225–D229. doi: 10.1093/nar/gkq1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang Y, Geer LY, Chappey C, Kans JA, Bryant SH. Cn3D: sequence and structure views for Entrez. Trends Biochem. Sci. 2000;25:300–302. doi: 10.1016/s0968-0004(00)01561-9. [DOI] [PubMed] [Google Scholar]
- 54.Gibrat JF, Madej T, Bryant SH. Surprising similarities in structure comparison. Curr. Opin. Struct. Biol. 1996;6:377–385. doi: 10.1016/s0959-440x(96)80058-3. [DOI] [PubMed] [Google Scholar]
- 55.Madej T, Gibrat JF, Bryant SH. Threading a database of protein cores. Proteins. 1995;23:356–369. doi: 10.1002/prot.340230309. [DOI] [PubMed] [Google Scholar]
- 56.Marchler-Bauer A, Anderson JB, Chitsaz F, Derbyshire MK, DeWeese-Scott C, Fong JH, Geer LY, Geer RC, Gonzales NR, Gwadz M, et al. CDD: specific functional annotation with the conserved domain database. Nucleic Acids Res. 2009;37:D205–D210. doi: 10.1093/nar/gkn845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Letunic I, Copley RR, Pils B, Pinkert S, Schultz J, Bork P. SMART 5: domains in the context of genomes and networks. Nucleic Acids Res. 2006;34:D257–D260. doi: 10.1093/nar/gkj079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Finn RD, Mistry J, Schuster-Bockler B, Griffiths-Jones S, Hollich V, Lassmann T, Moxon S, Marshall M, Khanna A, Durbin R, et al. Pfam: clans, web tools and services. Nucleic Acids Res. 2006;34:D247–D251. doi: 10.1093/nar/gkj149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Haft DH, Selengut JD, White O. The TIGRFAMs database of protein families. Nucleic Acids Res. 2003;31:371–373. doi: 10.1093/nar/gkg128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang Y, Xiao J, Suzek TO, Zhang J, Wang J, Bryant SH. PubChem: a public information system for analyzing bioactivities of small molecules. Nucleic Acids Res. 2009;37:W623–W633. doi: 10.1093/nar/gkp456. [DOI] [PMC free article] [PubMed] [Google Scholar]