Abstract
Saccharomyces cerevisiae is a primary model for studies of transcriptional control, and the specificities of most yeast transcription factors (TFs) have been determined by multiple methods. However, it is unclear which position weight matrices (PWMs) are most useful; for the roughly 200 TFs in yeast, there are over 1200 PWMs in the literature. To address this issue, we created ScerTF, a comprehensive database of 1226 motifs from 11 different sources. We identified a single matrix for each TF that best predicts in vivo data by benchmarking matrices against chromatin immunoprecipitation and TF deletion experiments. We also used in vivo data to optimize thresholds for identifying regulatory sites with each matrix. To correct for biases from different methods, we developed a strategy to combine matrices. These aligned matrices outperform the best available matrix for several TFs. We used the matrices to predict co-occurring regulatory elements in the genome and identified many known TF combinations. In addition, we predict new combinations and provide evidence of combinatorial regulation from gene expression data. The database is available through a web interface at http://ural.wustl.edu/ScerTF. The site allows users to search the database with a regulatory site or matrix to identify the TFs most likely to bind the input sequence.
INTRODUCTION
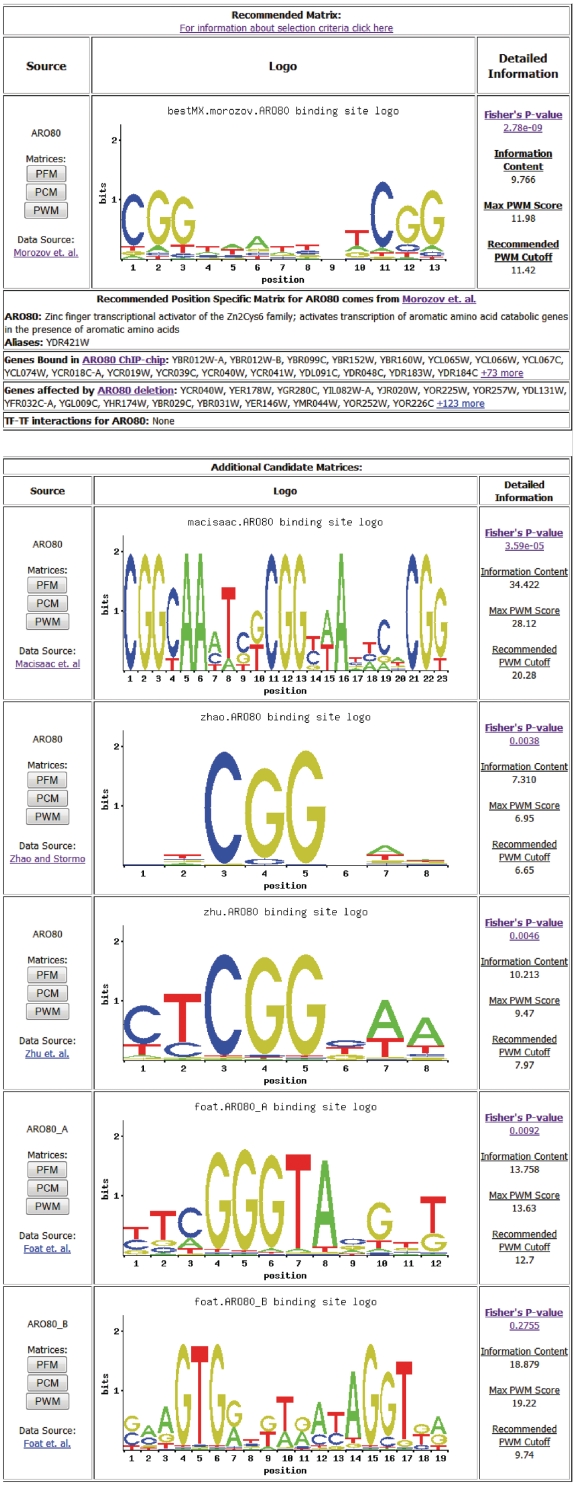
To understand gene regulation, it is necessary to accurately identify transcription factor (TF) binding sites in the genome. Over the past decade, numerous studies have been published that predict the DNA binding specificities of TFs in Saccharomyces cerevisiae (1–8). Each of these studies relied on different experimental and computational strategies to generate models of DNA–protein interactions in the form of position-specific weight matrices (PWMs) (9). Each of the different methods is subjected to different biases, which may produce accurate models of specificity for certain types of TFs but not others. Since the binding specificities of yeast TFs have been intensively studied, there are multiple, often conflicting, PWMs for most TFs (Figure 1). No existing database provides a comprehensive repository of available PWMs, and there has been no systematic effort to evaluate the predictive ability of each PWM using in vivo data sets as a benchmark. ScerTF provides a collection of matrices that are optimal, among the set of PWMs considered, for predicting in vivo TF occupancy.
Figure 1.
Screenshot of details page for ARO80. There are multiple, conflicting PWMs available from five different literature sources for ARO80. The details page provides P-values and recommended cutoffs for each PWM as well as target genes identified in ARO80 ChIP-chip (4) and gene deletion experiments (13).
Existing databases cover only a limited number of TFs or lack objective validation. The SwissRegulon database (7) is one repository of PWMs, but most models are derived only from phylogenetic footprinting and the database contains data for 72 TFs. The Saccharomyces cerevisiae promoter database (SCPD) (10) contains PWMs for just 24 factors. The most recent version of JASPAR (11) is, to date, the most complete collection, with results for 176 unique yeast TFs. The JASPAR curators collected PWMs from five different sources including SwissRegulon and SCPD, but prioritized the sources based on the curators’ personal perspectives. A matrix from a low-priority source was discarded if a high-priority source already contained a matrix annotated to the same TF. In many cases, the prioritized source was a collection of matrices produced by various in vitro binding assays (1). Such assays are high-throughput and generally reliable, but are not guaranteed to provide the most accurate representation of a TF's in vivo binding specificity (12). This is especially true for TFs that dimerize to bind DNA (1,12).
We created ScerTF, a comprehensive, curated database that incorporates PWMs derived from a variety of experimental and computational methods. The database contains 1226 matrices from 11 different sources, covering 196 different TFs. For each TF in the database, we evaluated the available matrices by comparing matrix-predicted TF binding sites against results from in vivo chromatin immunoprecipitation (ChIP) occupancy (4) and TF deletion (13) experiments. Based on this evaluation, we provide a compendium of the best-performing matrices and we also provide performance metrics for all matrices annotated to a particular TF (Figure 1). This allows the user to individually compare the recommended matrix with additional candidate matrices. Because transcription factors bind degenerate sets of sequences, we have also employed the ChIP-chip data to determine an optimal cutoff to use when searching for potential regulatory sites.
The two experimental methods we chose to use as benchmarks in this effort measure the in vivo activity of the transcriptional regulators assayed. ChIP measures the physical interaction between a protein or protein complex and DNA, providing a direct readout of binding locations in the genome. Expression analysis of a TF gene deletion mutant measures the direct and indirect genetic interactions between a transcriptional regulator and downstream target genes. The motivation of ScerTF is to provide scientists with a way to accurately identify TF binding sites in the genome. As such, comparing matrices with results from in vivo experimental data is a natural metric to use as a benchmark.
In addition to curating data sets from the literature, we also developed a strategy to optimize a PWM given a collection of matrices and applied this method to the TFs curated in the database. Our strategy was able to generate matrices that outperformed the best existing PWMs in predicting TF occupancy for ∼10% of the TFs in the database.
We demonstrate the use of ScerTF in two ways. First, we provide an example from the recent literature in which a group identified functional cis-regulatory elements (CREs) but were unable to associate these regulatory sites with specific TFs. By searching the ScerTF database with these published regulatory sequences, we were able to identify potential regulators that bind the appropriate target genes in ChIP experiments and significantly affect expression of the appropriate target genes when deleted. Second, we used the collection of motifs in the database to identify instances of combinatorial regulation in the yeast genome. This search identified many known CRE combinations and also predicts new combinations in which the target genes of the combination are coherently expressed.
DATABASE ASSEMBLY AND CURATION
To create ScerTF, we collected results from 11 different computational and experimental studies that report binding specificities of TFs in S. cerevisiae. These studies rely on different methods to infer DNA binding specificities, including phylogenetic footprinting (5,7), molecular modeling (6), gene expression analysis (2), in vitro binding assays (3), ChIP (4), DNA immunoprecipitation with microarray detection (DIP-ChIP) (1) and protein binding microarrays (1,8,14). In addition, we incorporated the SCPD database (10) into our own database to evaluate the performance of its matrices and to assimilate these matrices into our alignment strategy. Matrices from the commercially available TRANSFAC database were also evaluated using the same metrics, but are not made freely available in this database (15). However, in all the cases, the TRANSFAC PWMs were outperformed by matrices in at least one of the other data sets.
For the Badis et al. (1), Foat et al. (2), Morozov and Siggia (6), Zhu et al. (8) and Zhao and Stormo (14) data sets, the matrices generally have a core motif with high information content that is surrounded by uninformative flanking positions at the edges of the matrix. Following a recent method by Badis et al. (16), we trim the distal positions of these matrices based on information content. The distal columns of a matrix were removed if the information content of those columns dropped below 0.3. To standardize the naming system across the literature sources, we converted all matrix names to the common name for the TF provided by the Saccharomyces Genome Database (17). Matrix logos were generated using tools available from Lenhard and Wasserman (18).
To identify the best-performing matrices, we evaluated each matrix using in vivo experimental data. When available for a particular TF, we used ChIP-chip binding data from a compendium of experiments performed by Harbison et al. (4). To compare matrices, we first designated promoters as either bound or unbound according to the P-values published for each ChIP experiment. Following the method of Harbison et al. (4), we consider sequences with a P < 0.001 to be bound. For each ChIP experiment, the matrices annotated to that TF in ScerTF were used to predict bound probes by identifying the maximum score produced by that matrix for each probe sequence and designating the probe as either bound or unbound depending on whether that score exceeded a given threshold. Thus, to accurately predict in vivo regulatory sites, a matrix must not only identify binding sites in bound probes but also predict that unbound probes do not contain a binding site. These predictions were compared with the observed occupancy in the ChIP experiment using the Fisher's exact test (19). The matrix with the best P-value is marked as the recommended matrix in the database. We optimize the cutoff used to predict TF binding for each matrix by starting with a threshold of 50% of the consensus sequence PWM score and iteratively increasing the threshold so that the lowest scoring sequence transitions from the predicted ‘bound’ set to the predicted ‘unbound’ set. At each step, the Fisher's exact P-value is recalculated and the cutoff is optimized when the P-value is minimized. For several TFs, Harbison et al. (4) measured TF occupancy in more than one growth condition; in these cases, we consider each condition separately and implement a Bonferroni multiple hypothesis correction so that P-values are determined consistently for all factors independent of the number of conditions.
In the event that ChIP data were unavailable for a particular TF, matrices were evaluated using data obtained from an analysis of TF gene deletion mutants (13). In this data set, genes are annotated as either significantly up- or downregulated in response to the deletion of a particular TF. Each matrix annotated to a particular TF was used to predict which genes should be up/downregulated in a deletion mutant strain for that TF as described above. Predictions were compared with observed data using the Fisher's exact test. For 16 TFs, there was neither ChIP-chip nor TF deletion data available to help decide among multiple available PWMs; for these cases, we choose between matrices based on corroborating evidence from literature searches (Supplementary Table S1).
If we could identify corroborating evidence to support a particular matrix from experimental studies of individual TFs, we gave priority to that evidence over results from high-throughput analyses. For the TF ARR1, the highest performing matrix comes from Foat et al. (2) but this PWM is inconsistent with the other literature sources and an in-depth analysis of ARR1 (also called YAP8) by Wysocki et al. (20). The recommended ARR1 matrix reflects the experimental results from Wysocki et al. (20) and shares the same TTA core half-site with related YAP proteins. A similar situation arises for ROX1, in which the best-performing matrix does not match the canonical binding motif of the SOX protein family (21). However, a matrix from Fordyce et al. (3) significantly predicts ChIP binding data and reflects the binding specificities determined for ROX1 by detailed experimentation (22). Both FHL1 and SFP1 matrices derived from ChIP-chip data closely resemble the DNA binding specificity of RAP1 and are inconsistent with in vitro measurements (1,8). Because FHL1 and SFP1 may cooperatively bind DNA with RAP1 (23), their true binding specificities are masked in the ChIP-chip data, and therefore, the recommended matrices for these two TFs come from in vitro measurements. Similarly, the recommended matrix for YOX1 comes from in vitro data because YOX1 is known to cooperatively bind with MCM1 (24).
Some of the TFs included in ScerTF are not known to be sequence-specific DNA binding proteins and are likely the co-factors that interact with other TFs to bind the genome. However, it is important to include these factors in the database because they contribute to specific regulation of gene expression, and, as in the case of SWI6, they frequently exhibit sequence-specificity in their genomic location because they associate with sequence-specific factors that bind DNA directly. For TFs that are not associated with the Gene Ontology term for ‘sequence-specific DNA binding’ (25), like SWI6, UME1, THO2, SPT2 and SNT2, we have included a disclaimer in ScerTF that the motifs described for these proteins may actually represent the specificity of a cofactor. In these contexts, the predictive motif chosen in ScerTF does not reflect the DNA binding specificity of the given protein, but most accurately predicts where the factor will be complexed with DNA in vivo.
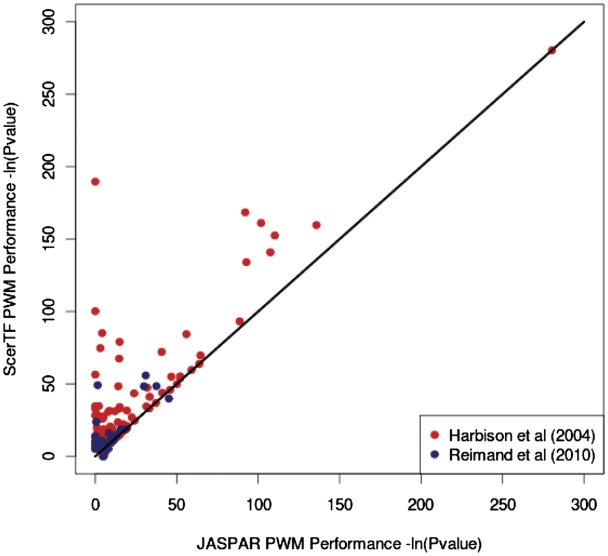
To evaluate our selection of recommended matrices, we compared the optimal ScerTF matrices to the JASPAR matrices using two different experimental data sets (4,13). PWMs from each database were benchmarked as described above, using the Fisher's exact test to assess the ability of a PWM to predict bound and unbound ChIP probes. The results of this analysis are largely consistent regardless of input data set. In almost all cases in which either database could significantly predict experimental data, the ScerTF recommended matrices outperform JASPAR matrices (Figure 2).
Figure 2.
Comparison of ScerTF recommended weight matrices with JASPAR weight matrices. Count matrices from JASPAR were converted to weight matrices as described in Ref. (9). Each matrix was used to predict which promoters should be bound by a TF and which promoters should not be bound. Predictions were compared with results from ChIP-chip experiments as reported by Harbison et al. (4) and gene deletion experiments as reported by Reimand et al. (13) using the Fisher's Exact test.
MATRIX ALIGNMENT
The individual studies incorporated into this database all employed different experimental and computational strategies to determine the DNA binding specificity of yeast TFs. Ideally, different methods should accurately parameterize DNA–protein interactions and thus produce identical PWMs for a given TF. In actuality, however, each method is vulnerable to different biases and ultimately produces an approximation of a TF's true specificity. To minimize the impact of artifacts introduced by individual methods, we devised a strategy to align and combine matrices annotated to the same TF across multiple studies for cases in which we could rely on experimental data to refine available matrices. We have implemented a ‘Glocal’ alignment strategy to identify a common motif in two position frequency matrices (PFMs), using the average log-likelihood ratio (26) as a measure of similarity between positions in the alignment. As in a local alignment, we do not penalize overhangs when aligning matrices. As in a global alignment, we require that a match must extend to the end of a matrix. Additionally, we do not allow gaps when aligning two matrices. Once an optimal alignment has been found, we combine the matrices by averaging the nucleotide frequencies at matched positions within the alignment; overhanging segments in the alignment that remain unmatched are averaged against the background nucleotide frequency of the S. cerevisiae genome. The flanking positions of this long, aligned matrix are then trimmed to eliminate positions with low-information content.
The alignment search is performed in two steps. First, each individual matrix from the different data sets is scored on its ability to predict bound and unbound sequences in ChIP experiments (4) using the Fisher's exact test. The matrices are ranked by P-value and then the second best matrix is aligned against the first. Next, the aligned matrices are combined and the resulting matrix is benchmarked and added to a new set of candidate matrices. Each additional matrix from the original set is aligned against all matrices with better performance, and the optimal alignment is then used to generate a new matrix. This new aligned matrix is ranked and added to the set of candidate matrices. The algorithm progresses through the original list of matrices until it is exhausted and then identifies the aligned matrix with the best performance. In most cases, the matrix produced from this procedure outperforms some, but not all of the original matrices. However, for ∼10% of all TFs in the database, the aligned matrix outperforms all of the matrices in the original data set (Figure 3).
Figure 3.
PWMs available for GCR1 in the literature vary in both information content and length. By combining the available matrices, we were able to produce a PWM for GCR1 that more accurately discriminates between bound and unbound probes in a GCR1 ChIP-chip experiment.
MATRIX AND NUCLEOTIDE SEQUENCE SEARCH
An essential feature of ScerTF is the ability to compare an input DNA sequence or PWM against the entire catalog of TF binding specificities. The search feature that we have implemented allows users to query the database with a single cis-regulatory site or with a consensus-formatted matrix (9). For each matrix in the database, we align the input sequence or matrix using the ‘Glocal’ alignment method described above. For the aligned portion of the database PWM, we calculate the mean and variance of PWM scores for all possible DNA sequences, which provides a distribution of all possible scores the aligned region can produce. If the user has submitted a DNA sequence, we calculate the deviation of the input sequence from the mean of this score distribution using a Z-test and additionally calculate the relative affinity of that sequence compared with the consensus sequence for the PWM alignment. If the user has submitted a matrix, we convert the input matrix to a PFM and calculate the dot product of the user-input PFM and the ScerTF PWM to calculate the average PWM score over all sequences that contributed to the PFM (assuming positions in the PFM are independent). Using this average PWM score, we then calculated the deviation from the PWM mean using the Z-test as well as the average relative affinity compared with the consensus sequence for the PWM. We implement a Bonferroni multiple hypothesis correction to account for the number of comparisons performed against the query sequence/matrix.
When presented with an input nucleotide sequence that is shorter than a database matrix, we first identify the best possible alignment between the nucleotide sequence and the database matrix and use this as the score for that matrix. The greatest use of the search feature is to identify candidate matrices that could bind a regulatory sequence when very little is known about the sequence, including the full length of the regulatory site. However, in these cases, the reported match is only a partial match to a matrix in the database, and the true significance can only be determined by the context of the putative regulatory site.
The sequence and matrix search capability extends the utility of our database beyond yeast research. Any researcher studying transcriptional regulation in any organism can search this database for candidate matrices that closely match a cis-regulatory sequence or matrix produced by a motif discovery program. Although the TFs in this database are from S. cerevisiae, a matched S. cerevisiae TF can be used to identify the appropriate homolog in another organism of interest. This strategy has been successfully employed in previous studies to transfer knowledge from a model organism to a less well-studied organism (27–29). Yeast has been a major model organism for studies of transcriptional regulation, so its set of well-characterized TFs can be a key source of information about the behavior of homologous TFs in other organisms.
WEB INTERFACE AND APPLICATION
The ScerTF database is available at http://ural.wustl.edu/ScerTF. From the main webpage, a user can directly search for a TF by name, enter a regulatory sequence to identify potential regulators or enter a position-specific matrix to identify the most similar weight matrix in the database (Figure 1). Additionally, the entire database can be browsed or downloaded, including recommended cutoffs, by selecting the appropriate link in the navigation bar. Once a user identifies a TF of interest, the database will display all of the PWMs annotated to that TF and provide information about combinatorial regulation with other factors, promoters bound in ChIP-chip experiments (4), and genes significantly affected by TF gene deletion experiments (13). Each entry in the database also provides data on PWM performance in predicting ChIP or gene deletion data as well as a recommended cutoff to use when scanning a sequence for potential regulatory sites (Figure 1).
To illustrate the utility of the website, we found an example from the recent literature in which a group identified specific regulatory sequences but were unable to associate these sequences with specific TFs. In an examination of the YJL212C promoter, Srikanth et al. (30) identified a CRE with the sequence CGCCACA but were unable to determine the regulator that bound this site. Searching for this sequence in ScerTF reports MET31p as the top match and further reports that YJL212C was significantly bound in ChIP experiments and that deletion of MET31 significantly affects YJL212C expression. Although a motif was available for MET31 from a ChIP study published before Srikanth et al.’s investigation (4), there was no convenient way to search for a potential regulator using a single sequence.
USING ScerTF MATRICES TO PREDICT COMBINATORIAL REGULATION
A salient feature of eukaryotic gene regulation is that transcription initiation at a particular gene is controlled by multiple TFs (31). Early investigations of combinatorial gene regulation indicated that CREs that interact often cluster together in the genome (32). More recent studies have found that CREs which cluster together in one genome will still cluster together in related species even when the target genes differ between species (33). To capitalize on this observation, we used the matrices compiled in ScerTF to identify CREs that cluster in multiple related Saccharomyces species (manuscript in preparation). For each CRE combination we identified by this method, we predicted target genes of the combination in the S. cerevisiae genome. Target genes were identified by scanning promoter regions, defined as 600 bp upstream of each gene (34), for instances of the CRE combination. We used the optimal cutoffs calculated for each matrix and identified instances where regulatory elements co-occurred within a 25-nt window as evidence of a CRE combination. Genes with an occurrence of both regulatory elements within 25 bp of each other were predicted to be targets of the combination, while genes with a binding site for only one of the two regulatory elements were predicted to be targets of each CRE acting in isolation. We then analyzed gene expression data to determine if the CRE combination target genes were more coherently expressed than would be expected by chance. Three expression data sets were used to determine if predicted target genes of both CREs were co-expressed across multiple cell cycle time points (35), environmental conditions (36) or gene deletion conditions (37). For each data set, the Pearson's correlation coefficient (PCC) was calculated between gene expression profiles for all pairs of predicted target genes, which produced a distribution of PCC values describing the expression profile similarities of the target genes. This distribution of PCC values for predicted targets of the CRE combination was compared with the distribution of PCC values calculated for expression profiles of the targets in which each CRE was predicted to act in isolation. Differences between the distributions were assessed using the Mann–Whitney–Wilcoxon test. With this approach, we recovered many known combinations of cooperatively-acting TFs, including combinations involved in regulating the cell cycle (38) (Figure 4). We also identified new CRE combinations for which the expression profiles of the target genes are significantly more coherent than the expression profiles of genes targeted by each CRE individually. This information is available for each TF in the ScerTF database; identified regulatory interactions are listed for each TF along with the co-expression evidence (described in Figure 4).
Figure 4.
Expression profiles of FKH2xMCM1 target genes are more correlated than target genes of MCM1 and FKH2 acting alone. The yellow line depicts the distribution of correlation coefficients calculated between gene expression profiles for each pair of target genes predicted to be regulated by FKH2 and MCM1. The black and purple lines relate the distribution of correlations for target genes predicted to be regulated by either FKH2 or MCM1 alone.
DISCUSSION
Accurate prediction of TF binding sites is crucial to understanding the regulatory logic of gene regulation. ScerTF catalogs over 1200 PWMs for 196 different TFs, making it the most comprehensive database of yeast TFs available. We evaluated these matrices using data from ChIP-chip and TF deletion experiments to identify the most accurate PWM for each TF. ScerTF provides an unbiased compilation of the most accurate PWMs along with an optimal cutoff to use for each PWM when searching for cis-regulatory sites.
Ultimately, the goal of this effort is to identify a matrix from the available literature that explains in vivo TF occupancy data and accurately reflects the binding specificity of a given TF. As such, the collection of recommended matrices provided in ScerTF has been optimized for predicting in vivo occupancy using currently available experimental data sets. Each data set has particular contingencies and potential biases, which means that when assayed by other technologies or in other environmental conditions, a different set of matrices may be preferred.
ScerTF's search capability makes the database useful for a wide range of problems, such as linking regulatory sites with TFs, identifying a TF based on a user-input matrix, finding the genes bound/regulated by a particular TF, and finding regulatory interactions between TFs. Since many TFs are thought to be conserved between distant organisms (39), the database possesses utility for researchers outside of yeast genetics as well.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Table S1.
FUNDING
Funding for open access charge: National Institutes of Health (R01 GM078222 and HG00249).
Conflict of interest statement. None declared.
REFERENCES
- 1.Badis G, Chan ET, van Bakel H, Pena-Castillo L, Tillo D, Tsui K, Carlson CD, Gossett AJ, Hasinoff MJ, Warren CL, et al. A library of yeast transcription factor motifs reveals a widespread function for Rsc3 in targeting nucleosome exclusion at promoters. Mol. Cell. 2008;32:878–887. doi: 10.1016/j.molcel.2008.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Foat BC, Tepper RG, Bussemaker HJ. TransfactomeDB: a resource for exploring the nucleotide sequence specificity and condition-specific regulatory activity of trans-acting factors. Nucleic Acids Res. 2008;36:D125–D131. doi: 10.1093/nar/gkm828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fordyce PM, Gerber D, Tran D, Zheng J, Li H, DeRisi JL, Quake SR. De novo identification and biophysical characterization of transcription-factor binding sites with microfluidic affinity analysis. Nat. Biotechnol. 2010;28:970–975. doi: 10.1038/nbt.1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Harbison CT, Gordon DB, Lee TI, Rinaldi NJ, Macisaac KD, Danford TW, Hannett NM, Tagne JB, Reynolds DB, Yoo J, et al. Transcriptional regulatory code of a eukaryotic genome. Nature. 2004;431:99–104. doi: 10.1038/nature02800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.MacIsaac KD, Wang T, Gordon DB, Gifford DK, Stormo GD, Fraenkel E. An improved map of conserved regulatory sites for Saccharomyces cerevisiae. BMC Bioinformatics. 2006;7:113. doi: 10.1186/1471-2105-7-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Morozov AV, Siggia ED. Connecting protein structure with predictions of regulatory sites. Proc. Natl Acad. Sci. USA. 2007;104:7068–7073. doi: 10.1073/pnas.0701356104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pachkov M, Erb I, Molina N, van Nimwegen E. SwissRegulon: a database of genome-wide annotations of regulatory sites. Nucleic Acids Res. 2007;35:D127–D131. doi: 10.1093/nar/gkl857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhu C, Byers KJ, McCord RP, Shi Z, Berger MF, Newburger DE, Saulrieta K, Smith Z, Shah MV, Radhakrishnan M, et al. High-resolution DNA-binding specificity analysis of yeast transcription factors. Genome Res. 2009;19:556–566. doi: 10.1101/gr.090233.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hertz GZ, Stormo GD. Identifying DNA and protein patterns with statistically significant alignments of multiple sequences. Bioinformatics. 1999;15:563–577. doi: 10.1093/bioinformatics/15.7.563. [DOI] [PubMed] [Google Scholar]
- 10.Zhu J, Zhang MQ. SCPD: a promoter database of the yeast Saccharomyces cerevisiae. Bioinformatics. 1999;15:607–611. doi: 10.1093/bioinformatics/15.7.607. [DOI] [PubMed] [Google Scholar]
- 11.Portales-Casamar E, Thongjuea S, Kwon AT, Arenillas D, Zhao X, Valen E, Yusuf D, Lenhard B, Wasserman WW, Sandelin A. JASPAR 2010: the greatly expanded open-access database of transcription factor binding profiles. Nucleic Acids Res. 2010;38:D105–D110. doi: 10.1093/nar/gkp950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Berger MF, Bulyk ML. Universal protein-binding microarrays for the comprehensive characterization of the DNA-binding specificities of transcription factors. Nature protocols. 2009;4:393–411. doi: 10.1038/nprot.2008.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reimand J, Vaquerizas JM, Todd AE, Vilo J, Luscombe NM. Comprehensive reanalysis of transcription factor knockout expression data in Saccharomyces cerevisiae reveals many new targets. Nucleic Acids Res. 2010;38:4768–4777. doi: 10.1093/nar/gkq232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhao Y, Stormo GD. Quantitative analysis demonstrates most transcription factors require only simple models of specificity. Nat. Biotechnol. 2011;29:480–483. doi: 10.1038/nbt.1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Matys V, Fricke E, Geffers R, Gossling E, Haubrock M, Hehl R, Hornischer K, Karas D, Kel AE, Kel-Margoulis OV, et al. TRANSFAC: transcriptional regulation, from patterns to profiles. Nucleic Acids Res. 2003;31:374–378. doi: 10.1093/nar/gkg108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Badis G, Berger MF, Philippakis AA, Talukder S, Gehrke AR, Jaeger SA, Chan ET, Metzler G, Vedenko A, Chen X, et al. Diversity and complexity in DNA recognition by transcription factors. Science. 2009;324:1720–1723. doi: 10.1126/science.1162327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Engel SR, Balakrishnan R, Binkley G, Christie KR, Costanzo MC, Dwight SS, Fisk DG, Hirschman JE, Hitz BC, Hong EL, et al. Saccharomyces Genome Database provides mutant phenotype data. Nucleic Acids Res. 2010;38:D433–D436. doi: 10.1093/nar/gkp917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lenhard B, Wasserman WW. TFBS: Computational framework for transcription factor binding site analysis. Bioinformatics. 2002;18:1135–1136. doi: 10.1093/bioinformatics/18.8.1135. [DOI] [PubMed] [Google Scholar]
- 19.Marstrand TT, Frellsen J, Moltke I, Thiim M, Valen E, Retelska D, Krogh A. Asap: a framework for over-representation statistics for transcription factor binding sites. PLoS One. 2008;3:e1623. doi: 10.1371/journal.pone.0001623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wysocki R, Fortier PK, Maciaszczyk E, Thorsen M, Leduc A, Odhagen A, Owsianik G, Ulaszewski S, Ramotar D, Tamas MJ. Transcriptional activation of metalloid tolerance genes in Saccharomyces cerevisiae requires the AP-1-like proteins Yap1p and Yap8p. Mol. Biol. Cell. 2004;15:2049–2060. doi: 10.1091/mbc.E03-04-0236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mertin S, McDowall SG, Harley VR. The DNA-binding specificity of SOX9 and other SOX proteins. Nucleic Acids Res. 1999;27:1359–1364. doi: 10.1093/nar/27.5.1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Balasubramanian B, Lowry CV, Zitomer RS. The Rox1 repressor of the Saccharomyces cerevisiae hypoxic genes is a specific DNA-binding protein with a high-mobility-group motif. Mol. Cell Biol. 1993;13:6071–6078. doi: 10.1128/mcb.13.10.6071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gordan R, Hartemink AJ, Bulyk ML. Distinguishing direct versus indirect transcription factor-DNA interactions. Genome Res. 2009;19:2090–2100. doi: 10.1101/gr.094144.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pramila T, Miles S, GuhaThakurta D, Jemiolo D, Breeden LL. Conserved homeodomain proteins interact with MADS box protein Mcm1 to restrict ECB-dependent transcription to the M/G1 phase of the cell cycle. Genes Dev. 2002;16:3034–3045. doi: 10.1101/gad.1034302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gene Ontology Consortium. The Gene Ontology in 2010: extensions and refinements. Nucleic Acids Res. 2010;38:D331–D335. doi: 10.1093/nar/gkp1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang T, Stormo GD. Combining phylogenetic data with co-regulated genes to identify regulatory motifs. Bioinformatics. 2003;19:2369–2380. doi: 10.1093/bioinformatics/btg329. [DOI] [PubMed] [Google Scholar]
- 27.Hope IA, Struhl K. GCN4, a eukaryotic transcriptional activator protein, binds as a dimer to target DNA. EMBO J. 1987;6:2781–2784. doi: 10.1002/j.1460-2075.1987.tb02573.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yu J, Madison JM, Mundlos S, Winston F, Olsen BR. Characterization of a human homologue of the Saccharomyces cerevisiae transcription factor spt3 (SUPT3H) Genomics. 1998;53:90–96. doi: 10.1006/geno.1998.5500. [DOI] [PubMed] [Google Scholar]
- 29.Chodosh LA, Olesen J, Hahn S, Baldwin AS, Guarente L, Sharp PA. A yeast and a human CCAAT-binding protein have heterologous subunits that are functionally interchangeable. Cell. 1988;53:25–35. doi: 10.1016/0092-8674(88)90484-9. [DOI] [PubMed] [Google Scholar]
- 30.Srikanth CV, Vats P, Bourbouloux A, Delrot S, Bachhawat AK. Multiple cis-regulatory elements and the yeast sulphur regulatory network are required for the regulation of the yeast glutathione transporter, Hgt1p. Current genetics. 2005;47:345–358. doi: 10.1007/s00294-005-0571-7. [DOI] [PubMed] [Google Scholar]
- 31.Fickett JW, Wasserman WW. Discovery and modeling of transcriptional regulatory regions. Curr. Opin. Biotechnol. 2000;11:19–24. doi: 10.1016/s0958-1669(99)00049-x. [DOI] [PubMed] [Google Scholar]
- 32.Pilpel Y, Sudarsanam P, Church GM. Identifying regulatory networks by combinatorial analysis of promoter elements. Nat. Genet. 2001;29:153–159. doi: 10.1038/ng724. [DOI] [PubMed] [Google Scholar]
- 33.Tuch BB, Galgoczy DJ, Hernday AD, Li H, Johnson AD. The evolution of combinatorial gene regulation in fungi. PLoS Biol. 2008;6:e38. doi: 10.1371/journal.pbio.0060038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Singh LN, Hannenhalli S. Correlated changes between regulatory cis elements and condition-specific expression in paralogous gene families. Nucleic Acids Res. 2010;38:738–749. doi: 10.1093/nar/gkp989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pramila T, Wu W, Miles S, Noble WS, Breeden LL. The Forkhead transcription factor Hcm1 regulates chromosome segregation genes and fills the S-phase gap in the transcriptional circuitry of the cell cycle. Genes Dev. 2006;20:2266–2278. doi: 10.1101/gad.1450606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gasch AP, Spellman PT, Kao CM, Carmel-Harel O, Eisen MB, Storz G, Botstein D, Brown PO. Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell. 2000;11:4241–4257. doi: 10.1091/mbc.11.12.4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hughes TR, Marton MJ, Jones AR, Roberts CJ, Stoughton R, Armour CD, Bennett HA, Coffey E, Dai H, He YD, et al. Functional discovery via a compendium of expression profiles. Cell. 2000;102:109–126. doi: 10.1016/s0092-8674(00)00015-5. [DOI] [PubMed] [Google Scholar]
- 38.Bahler J. Cell-cycle control of gene expression in budding and fission yeast. Annu. Rev. Genet. 2005;39:69–94. doi: 10.1146/annurev.genet.39.110304.095808. [DOI] [PubMed] [Google Scholar]
- 39.Prud'homme B, Gompel N, Carroll SB. Emerging principles of regulatory evolution. Proc. Natl Acad. Sci. USA. 2007;104(Suppl. 1):8605–8612. doi: 10.1073/pnas.0700488104. [DOI] [PMC free article] [PubMed] [Google Scholar]