Abstract
The European Nucleotide Archive (ENA; http://www.ebi.ac.uk/ena), Europe's primary nucleotide sequence resource, captures and presents globally comprehensive nucleic acid sequence and associated information. Covering the spectrum from raw data to assembled and functionally annotated genomes, the ENA has witnessed a dramatic growth resulting from advances in sequencing technology and ever broadening application of the methodology. During 2011, we have continued to operate and extend the broad range of ENA services. In particular, we have released major new functionality in our interactive web submission system, Webin, through developments in template-based submissions for annotated sequences and support for raw next-generation sequence read submissions.
INTRODUCTION
The European Nucleotide Archive (ENA) is maintained and developed at the European Molecular Biology Laboratory's European Bioinformatics Institute (EMBL-EBI) and serves as Europe's primary repository for nucleotide sequence and associated information. Content spans raw sequence reads from all sequencing platforms, read alignments, assembly information and submitted functional annotation. Providing both the permanent scientific record as a complement to literature publication process and a forum for early sharing of pre-publication data, the ENA serves as a critical foundation for the global bioinformatics data infrastructure. Globally comprehensive coverage is assured through long-standing data exchange agreements with the DNA Databank of Japan (DDBJ) (1) and the United States National Institutes of Health National Center for Biotechnology Information (NCBI) (2) under the International Nucleotide Sequence Database Collaboration (3; http://www.insdc.org/).
Underlying ENA are a number of core databases, including the Sequence Read Archive for raw reads and read alignments from next generation sequencing platforms (4) and EMBL-Bank for high level assembly information, assembled sequences and functional annotation. ENA services are numerous: we provide submission tools, both the web-based Webin system and programmatic interfaces; we offer search technologies, such as the newly developed rapid ENA sequence similarity search (http://www.ebi.ac.uk/ena/search) and text-based search tools (http://www.ebi.ac.uk/ena); we present integrated access to all ENA content through the ENA Browser, which offers both web browsing and REST access (http://www.ebi.ac.uk/ena/about/browser). We are highly responsive in the development of new technologies and services to adapt to changes in sequencing technology and user requirements: we are leading a community-facing sequence read compression initiative, CRAM (5; http://www.ebi.ac.uk/ena/about/cram_toolkit); we are developing anencrypted BAM read alignment server that supports reference coordinate-based lookups of controlled acess reads by region; we are active in the development of data warehousing methodologies to provide real-time access to the massive data sets that we store (e.g. the ENA Taxon Portal; http://www.ebi.ac.uk/ena/data/view/Taxon:Eukaryota).
In this article, we comment on content and report briefly on means by which ENA data can be accessed. We then focus on major developments in our Webin submission system in the areas of template-based submissions of annotated and assembled sequences and raw next generation sequence read submission. We also announce the introduction of a sequence length limit for submission of assembled sequences.
ENA CONTENT
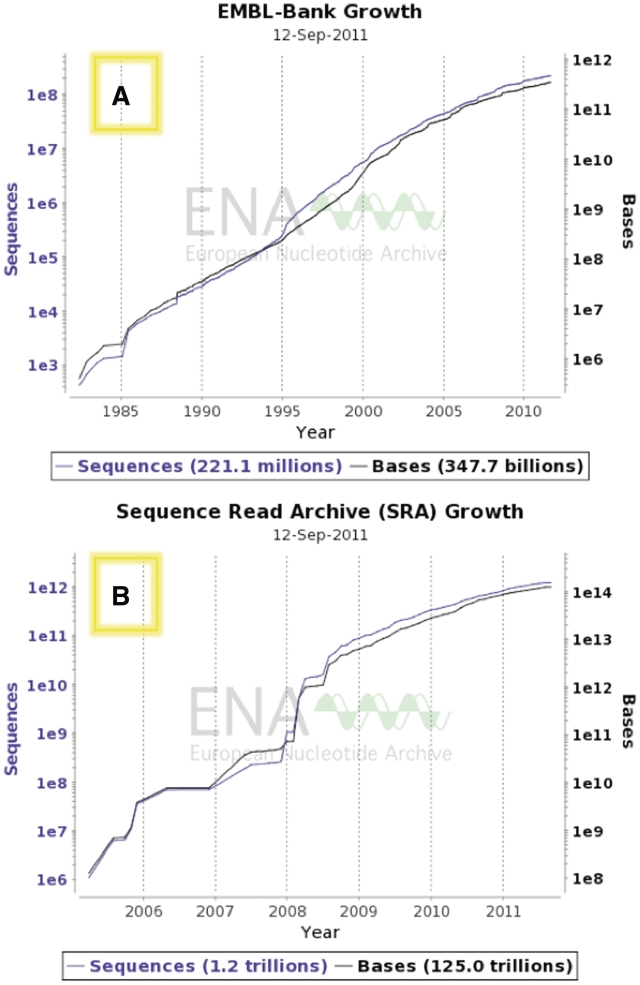
At the time of going to press, ENA contains 346 598 699 035 nt of assembled sequence in 220 504 007 assembled sequence entries (See EMBL-Bank release notes at http://www.ebi.ac.uk/embl/Documentation/Release_notes/current/relnotes.html) and more than 100 terabases of raw next generation sequence reads (Figure 1A and B).
Figure 1.
(A) Growth of assembled sequences (ENA:EMBL-Bank); see http://www.ebi.ac.uk/ena/about/statistics#embl_growth for dynamically updated growth chart. (B) Growth of raw data from next generation sequencing platforms (ENA: SRA); see http://www.ebi.ac.uk/ena/about/statistics#sra_growth for dynamically updated growth chart.
Notable datasets submitted to ENA during 2011 include assemblies of Gorilla gorilla (FR853080-FR853106), atlantic cod, Gadus morhua (Project:41391), Vine, Vitis vinifera (Project:18785), Takifugu rubripes (Project:1434), Macaca fascicularis (FR874244-FR874264), medieval mitochondria and Yersinia plasmids (6; HE576978-HE576987), raw genomic reads from 18 lines of Arabidopsis thaliana (7; ERP000565), Staphylococcus aureus (8; ERP000528) and Mus musculus ES cells (9; ERP000570) and transcriptomicreads from multiple Silene species (10; ERP000371).
ENA DATA ACCESS
Full ENA content is made available through an integrated platform, the ENA Browser, that supports discovery (text search, sequence similarity search, taxon lookup, etc.) and retrieval of records interactively (through web browsing and programatically under RESTful URLs). Full details are available from http://www.ebi.ac.uk/ena/about/browser. Records are made available in a selection of appropriate formats that include EMBL-Bank flat file, fasta and XML for assembled and annotated sequences, Fastq for sequence reads and Darwin Core for taxon records (http://www.ebi.ac.uk/ena/about/formats). In addition, we support both ftp and Aspera protocols for network transfers of large raw data sets (ftp://ftp.sra.ebi.ac.uk) and offer a variety of data products over ftp for other areas of ENA content (ftp://ftp.ebi.ac.uk/pub/databases/embl and ftp://ftp.ebi.ac.uk/pub/databases/ena)
ANNOTATED AND ASSEMBLED SEQUENCESUBMISSIONS
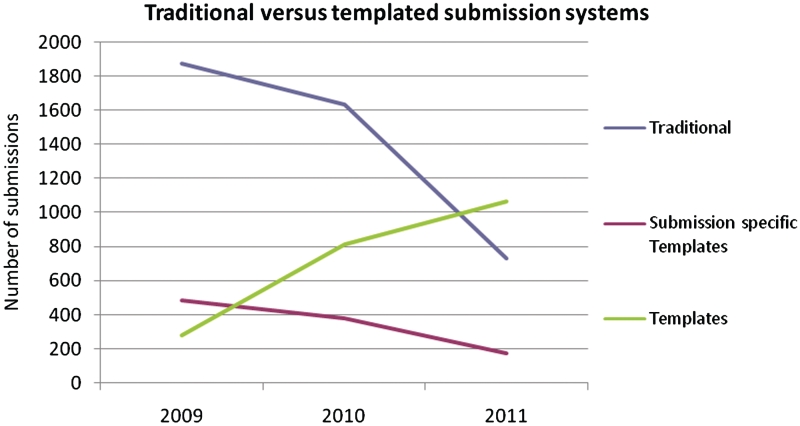
Apre-tailored template system was introduced in our Webin submission framework in 2009 for annotated sequence submissions and has been expanded during 2011 with the release of nine new templates. These templates have been designed for the most frequent types of sequence submissions and reached 15 in number in September 2011. When using the templates, submitters provide nucleotide sequences with associated annotation through spread sheets or Fastq files with pre-defined mandatory and optional fields, a process that significantly reduces the overall complexity of the submissions process for both the submitter and the ENA curator. Some advantages of the new system include the ability to choose from a small number of variables, functionalities that prevent the need for repetitive entry of information constant across all records in a data set and straightforward validation before data submission. The template concept has shown growing popularity since its launch versus the traditional system (which remains available for a limited time). Under the traditional system, submitters were able to annotate their entries with the full INSDC-approved features and qualifiers either one entry at a time or by defining with an ENA curator a specific template for each submission. This was useful for annotating small submissions in great detail but did not cater efficiently for larger-scale submissions of same-type data. Figure 2 shows the usage of the available submission systems between 2009 and 2011 and Table 1 shows the currently available templates.
Figure 2.
Usage of the different web-based interactive submission systems for annotated sequences at ENA between 2009 and 2011.
Table 1.
Names and definitions of templates currently available for sequence submissions to EMBL-bank
| Template name | Definition |
|---|---|
| Intergenic Spacer, IGS | For intergenic spacer (IGS) sequences between neighbouring genes (e.g. psbA-trnH IGS, 16S-23S rRNA IGS). Inclusion of the flanking genes is allowed |
| ITS region | For the 18S rRNA, ITS1, 5.8S rRNA, ITS2, 28S rRNA region, where the locations of the boundaries are not known |
| D-Loop | For mitochondrial D-loop (control region) sequences. All D-loops are considered partial |
| trnK-matK locus | For complete or partial matK gene within the chloroplast trnK gene |
| COI gene | For mitochondrial cytochrome oxidase subunit 1 genes |
| MHC gene 1 exon | For partial MHC class I or II antigens containing one exon |
| MHC gene 2 exons | For partial MHC class I or II antigens containing two exons |
| Single CDS genomic DNA | For complete or partial single non-segmented coding sequence (CDS) derived from genomic DNA |
| Single viral CDS genomic RNA | For complete or partial single coding sequence (CDS) derived from viral genomic RNA. Please do not use for viral DNA, peptides processed from polyproteins, viral cRNAs, or proviral sequences, as these are all annotated differently |
| Single CDS mRNA | For complete or partial single coding sequence (CDS) derived from mRNA (via cDNA) |
| rRNA gene | For ribosomal RNA genes from prokaryotic, nuclear or mitochondrial DNA. All rRNAs are considered partial |
| EST | For EST (expressed sequence tag) submissions |
| WGS (unannotated) | For unannotated Whole Genome Shotgun (WGS) sequences |
| MIMARKS-Survey 16S rRNA sequences | For the submission of 16S rRNA sequence compliant with the MIMARKS Minimal Information about a MARKer gene Sequence Standard |
| Soil sample MIMARKS-Survey using 16S rRNA sequences | For the submission of 16S rRNA sequence compliant with the MIMARKS Minimal Information about a MARKer gene Sequence Standard, specific to soil metagenomes |
As part of these developments, ENA is also facilitating the submission of marker gene sequences compliant with a community standard that has been developed by the Genomic Standards Consortium (GSC), called the Minimal Information about a MARKer gene Sequence Standard (MIMARKS) (11, 12). MIMARKS provides a minimal set of required information fields essential for downstream reuse of the data. The last two templates in Table 1 have been designed for submissions of MIMARKS-compliant data.
Further improvements to the submissions system for annotated sequences will continue in 2012 and beyond.
NEXT GENERATION SEQUENCE DATA SUBMISSIONS
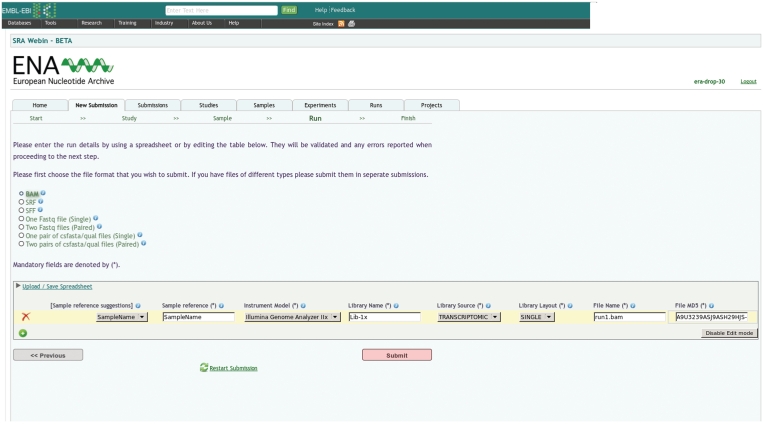
To complement the existing programmatic SRA REST submission interface, we have recently extended the Webin system to support submissions of raw next generation sequencing reads to the SRA. Unlike the SRA REST interface, which is targeted for large-scale sequence submitters and allows direct programmatic interaction between external LIMS systems and the SRA database at EBI, this new component of Webin is designed for interactive use. Users work through a web interface to create studies, samples and experiments, to update submitted metadata and to release previously submitted data to the public. Importantly, all metadata are submitted either by uploading or editing spreadsheets. While SRA REST submitters are fully exposed to the underlying SRA XML-data model, the SRA submission functionality in Webin completely hides this complexity. For example, during a raw sequence submission process, users are asked to define their raw data file format and are then presented with a spreadsheet, which can be either uploaded or filled with the required additional information (Figure 3).
Figure 3.
Screenshot of raw data definition page in SRA Webin.
The SRA submission component of Webin is under active development and new improvements are deployed weekly. Forthcoming improvements include support for European Genome–Phenome Archive submissions for controlled access raw sequence data, support for checklist for provision of community standard compliant meta data and numerous usability additions.
INTRODUCTION OF SEQUENCE LENGTH LIMIT FOR ASSEMBLED SEQUENCES
ENA will introduce a sequence length limit for submissions of assembled sequences. From January 2012, ENA will accept sequences <100 bp only if they fall into one of the following sequence categories of ‘Ancient DNA’, ‘non-coding-RNA’, ‘Microsatellites’ or ‘Complete Exons’. Exceptions require the submitter to demonstrate that a peer-reviewed journal has accepted a manuscript by the submitter, confirming the relevance of the short sequences to the scientific community. A validation step will be implemented in Webin to facilitate implementation of this requirement. We encourage submitters to check our website for further forthcoming changes announcements (http://www.ebi.ac.uk/ena/about/forthcoming_changes)
HELPDESK AND TRAINING
The ENA team provides advice and guidance regarding ENA services by email through datasubs@ebi.ac.uk. Feedback and suggestions related to all of our services are very welcome at the same email address. We also operate a variety of hands-on training programmes, for which details are available at http://www.ebi.ac.uk/training. We strongly encourage submitters to take our survey (http://www.surveymonkey.com/s/ENA_User_Survey_2011) and help us to improve our service.
FUNDING
European Molecular Biology Laboratory; FP7 Programme of the European Commission; WellcomeTrust and Biotechnology and Biological Sciences Research Council (BBSRC). Funding for open access charge: European Molecular Biology Laboratory.
Conflict of interest statement. None declared.
REFERENCES
- 1.Kaminuma E, Kosuge T, Kodama Y, Aono H, Mashima J, Gojobori T, Sugawara H, Ogasawara O, Takagi T, Okubo K, et al. DDBJ progress report. Nucleic Acids Res. 2011;39:D22–D27. doi: 10.1093/nar/gkq1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW. Genbank. Nucleic Acids Res. 2011;39:D32–D37. doi: 10.1093/nar/gkq1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cochrane G, Karsch-Mizracthi I, Nakamura Y, International Nucleotide Sequence Database Collaboration. (2011) The international nucleotide sequence database collaboration in 2010. Nucleic Acids Res. 39:D15–D18. doi: 10.1093/nar/gkq1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Leinonen R, Sugawara H, Shumway M, International Nucleotide Sequence Database Collaboration The sequence read archive. Nucleic Acids Res. 2011;39:D19–D21. doi: 10.1093/nar/gkq1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hsi-Yang Fritz M, Leinonen R, Cochrane G, Birney E. Efficient storage of high throughput DNA sequencing data using reference-based compression. Genome Res. 2011;21:734–740. doi: 10.1101/gr.114819.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schuenemann VJ, Bos KI, DeWitte SN, Jamieson J, Mittnik A, Forrest SA, Coombes BK, Wood JW, Earn D, White W, et al. Targeted enrichment of ancient pathogens yielding the pPCP1 plasmid of Yersinia pestis from victims of the Black Death. Proc. Natl Acad. Sci., USA. 2011;108:E746–E752. doi: 10.1073/pnas.1105107108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gan X, Stegle O, Behr J, Steffen JG, Drewe P, Hildebrand KL, Lyngsoe R, Schultheiss SJ, Osborne EJ, Sreedharan VT, et al. Multiple reference genomes and transcriptomes for Arabidopsis thaliana. Nature. 2011;477:419–423. doi: 10.1038/nature10414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Corrigan RM, Abbott JC, Burhenne H, Kaever V, Grundling A. -di-AMP is a new second messenger in Staphylococcus aureus with a role in controlling cell size and envelope stress. PLoS Pathog. 2011;7:e1002217. doi: 10.1371/journal.ppat.1002217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ficz G, Branco MR, Seisenberger S, Santos F, Krueger F, Hore TA, Marques CJ, Andrews S, Reik W. Dynamic regulation of 5-hydroxymethylcytosine in mouse ES cells and during differentiation. Nature. 2011;473:398–402. doi: 10.1038/nature10008. [DOI] [PubMed] [Google Scholar]
- 10.Blavet N, Charif D, Oger-Desfeux C, Marais GA, Widmer A. Comparative high-throughput transcriptome sequencing and development of SiEST, the Silene EST annotation database. BMC Genomics. 2011;12:376. doi: 10.1186/1471-2164-12-376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yilmaz P, Kottmann R, Field D, Knight R, Cole JR, Amaral-Zettler L, Gilbert JA, Karsch-Mizrachi I, Johnston A, Cochrane G, et al. Minimum information about a marker gene sequence (MIMARKS) and minimum information about any (x) sequence (MIxS) specifications. Nat. Biotechnol. 2011;29:415–420. doi: 10.1038/nbt.1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yilmaz P, Gilbert JA, Knight R, Amaral-Zettler L, Karsch-Mizrachi I, Cochrane G, Nakamura Y, Sansone SA, Glöckner FO, Field D. The genomic standards consortium: bringing standards to life for microbial ecology. ISME J. 2011;5:1565–1567. doi: 10.1038/ismej.2011.39. [DOI] [PMC free article] [PubMed] [Google Scholar]