Abstract
GeneSigDB (http://www.genesigdb.org or http://compbio.dfci.harvard.edu/genesigdb/) is a database of gene signatures that have been extracted and manually curated from the published literature. It provides a standardized resource of published prognostic, diagnostic and other gene signatures of cancer and related disease to the community so they can compare the predictive power of gene signatures or use these in gene set enrichment analysis. Since GeneSigDB release 1.0, we have expanded from 575 to 3515 gene signatures, which were collected and transcribed from 1604 published articles largely focused on gene expression in cancer, stem cells, immune cells, development and lung disease. We have made substantial upgrades to the GeneSigDB website to improve accessibility and usability, including adding a tag cloud browse function, facetted navigation and a ‘basket’ feature to store genes or gene signatures of interest. Users can analyze GeneSigDB gene signatures, or upload their own gene list, to identify gene signatures with significant gene overlap and results can be viewed on a dynamic editable heatmap that can be downloaded as a publication quality image. All data in GeneSigDB can be downloaded in numerous formats including .gmt file format for gene set enrichment analysis or as a R/Bioconductor data file. GeneSigDB is available from http://www.genesigdb.org.
INTRODUCTION
Accurately curated and annotated gene sets have emerged as essential tools for the analysis of large, complex biological datasets. Gene set analysis (GSA) is widely used in the analysis and interpretation of gene expression profiling data (1–4), evolutionary relationships (5), genomic associations—including QTL analysis (6), genotyping (7) and SNP chips (8)—and even for cross platform integration of genomics data (9). GSA aims to find sets of genes that collectively distinguish two phenotypes, even if the genes in the set are not significantly different when tested individually. This reflects the fact that genes within the cell function as members of complex networks and pathways, often with multiple, overlapping functions. As a result, direct comparisons of genes may miss biologically important connections that are only seen when these related genes are assessed collectively.
Gene sets have also become invaluable tools for characterizing and distinguishing phenotypic states. In breast cancer, for example, several gene expression signatures have been developed as commercial diagnostic assays (10) and new methods are being developed that combine the predictive strength of multiple gene signatures to increase their prognostic power (11).
Gene set resources can be broadly divided into those which assign a gene to collections based on ‘known’ gene or protein interactions or functional activity and those that include gene lists from high-throughput experimental assays. Functional and pathway databases such as Gene Ontology (GO), KEGG and Reactome capture published descriptions of cellular pathways and gene functions (12), including, in the case of GO, functional predictions inferred from orthologous sequences (13). However, these resources are incomplete as we have not yet been able to comprehensively and completely catalog the functions of all genes in the genome (13).
High-throughput experiments, such as microarray expression profiling and RNA-seq have also produced large numbers of potentially informative gene lists. Most genomics papers present one or more gene signatures that reportedly correlate with experimental phenotypes. While there has been some controversy over the value of individual gene sets, due to the fact that many fail to fully replicate in independent data sets, the analysis of the collected gene lists defined for similar phenotypes has been demonstrated to provide meaningful biological insight (14).
Despite tremendous interest in using gene signatures, public repositories such as GEO and ArrayExpress (15,16) store primary gene expression data but fail to capture the gene sets that are the end product of published analyses. Without a systematic way of reporting these, the gene sets often appear only in published tables or figures or in supplementary materials hosted on the author's or the journal's website. And as there are no accepted standards for reporting gene sets, they often appear with non-standard gene identifiers, making comparison to other lists, or even to the original data, a significant challenge. Because of these limitations, gene sets from published research studies are often inaccessible to automated computational analysis.
In August 2009, we created GeneSigDB (17) as a repository for gene sets that had been systematically collected and manually curated from published articles indexed by PubMed. Our approach in building GeneSigDB was to capture gene signatures from the literature as published, to map them to standard identifiers using transparent, reproducible protocols and to freely provide these to the research community together with some elementary analytical tools. Since its launch, GeneSigDB had 7918 web hits with 4404 hits in 2010 and 3354 so far this year, suggesting that this resource is of value to the biomedical research community.
GROWTH OF GeneSigDB DATABASE
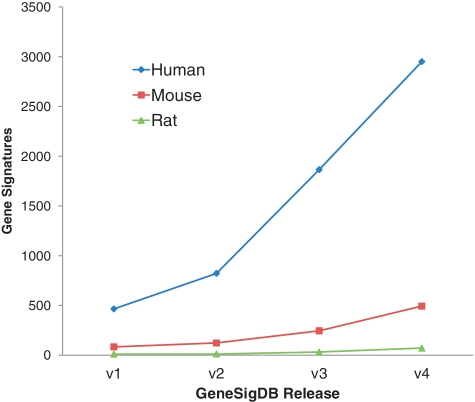
GeneSigDB has grown considerably since its introduction (Figure 1), nearly doubling in size with each subsequent release. GeneSigDB 4.0, released in September 2011, contains 3515 human, mouse and rat gene sets curated from 1604 published articles. While we have continued to focus on gene sets related to cancer and stem cells, we now also include signatures for development, inflammation and immune regulation, and lung disease and we have begun to catalog signatures for miRNA expression and proteomics. The content of GeneSigDB and its composition are summarized in Tables 1 and 2.
Figure 1.
Growth of GeneSigDB. GeneSigDB has grown considerably over 4 database releases (August 2009, March 2010, December 2010, September 2011). The most recent release (Release 4.0, September 2011) contains 3515 human, mouse and rat gene sets curated from 1604 published articles.
Table 1.
Number of processed articles and extracted gene signatures (by species) in GeneSigDB
| Human | Mouse | Rat | Total | |
|---|---|---|---|---|
| Gene Signatures | 2951 | 493 | 71 | 3515 |
| Publications (PMIDs) | 1368 | 208 | 39 | 1604* |
| Genes (EnsEMBL gene IDs) | 20 478 | 16 009 | 5110 |
*There were 10 articles with human and mouse gene signatures, and 1 article with human and rat gene signatures.
Table 2.
Most common disease MeSH terms associated with articles in GeneSigDBa
| MeSH Terms | Publications |
|---|---|
| Breast neoplasms | 248 |
| Lung neoplasms | 97 |
| Prostatic neoplasms | 73 |
| Disease progression | 69 |
| Neoplasm metastasis | 66 |
| Ovarian neoplasms | 66 |
| Adenocarcinoma | 65 |
| Cell transformation, neoplastic | 62 |
| Neoplasm invasiveness | 62 |
| Carcinoma, squamous cell | 58 |
| Liver neoplasms | 56 |
| Carcinoma, hepatocellular | 51 |
| Lymphatic metastasis | 42 |
| Colonic neoplasms | 38 |
| Neoplasms | 37 |
| Precursor cell lymphoblastic leukemia–lymphoma | 37 |
| Stomach neoplasms | 35 |
| Neovascularization, pathologic | 34 |
| Disease models, animal | 33 |
| Genetic predisposition to disease | 32 |
| Pancreatic neoplasms | 30 |
| Chromosome aberrations | 29 |
| Carcinoma | 28 |
| Leukemia, myeloid, acute | 28 |
| Brain neoplasms | 27 |
| Carcinoma, non-small-cell lung | 27 |
| Leukemia, myeloid | 25 |
| Neoplasm recurrence, local | 25 |
| Leukemia, lymphocytic, chronic, B-cell | 24 |
aRanking of Diseases MeSH Terms (MeSH prefix code category C) associated with 1552 publications in GeneSigDB. A total of 63 publications were not annotated with MeSH Terms. More details are provided in documentation on the GeneSigDB website.
GeneSigDB had minimal overlap with other gene signatures resources when we compared the overlap of publications curated by MSigDB (18) and CCancer (19) to GeneSigDB. Only 198/1604 (12%) publications were curated by both GeneSigDB (n = 1604) and MSigDB (n = 786, Release 3.0). GeneSigDB and MSigDB are both manually curated, but CCancer gene lists are computationally extracted from publications in 100 journals indexed by PubMed (19), and ∼30% of publications in CCancer are also manually curated in GeneSigDB. To estimate of curation quality, we examined 121 publications that were curated by all three gene signatures resources. The number of gene signatures identified in these 121 publications were 428 287 and 123 by MSigDB, GeneSigDB and CCancer respectively. MSigDB and GeneSigDB captured more data per publication that the automated curation of CCancer.
GENE SIGNATURE COLLECTION AND CURATION PIPELINE
The primary data objects in GeneSigDB are genes, published articles and gene signatures from those publications. We define a gene signature as a set of gene identifiers that were experimentally derived from analysis of gene, protein or miRNA expression.
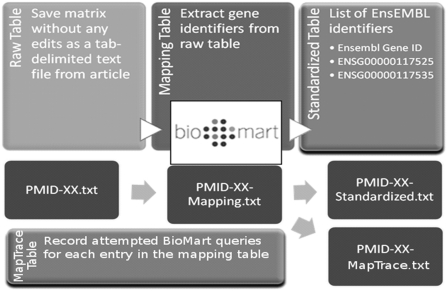
Published articles likely to contain gene signature are identified using predefined PubMed searches as described at http://compbio.dfci.harvard.edu/genesigdb/documentation.jsp. We download and read each article, identify tables or figures containing gene signatures, and transcribe these from the main body of the article, its supplementary materials or websites referenced within the article (Figure 2). We manually transcribe the entire table and then use a pipeline based on Biomart (20) to map the published gene identifiers to EnsEMBL IDs, creating standardized gene sets.
Figure 2.
Overview of the GeneSigDB Data Curation pipeline. Gene signatures in tables or figures are transcribed from published articles indexed in PubMed and we then use a pipeline based on Biomart (20) to map all published gene identifiers to EnsEMBL IDs, to create standardized gene sets.
The number of genes in the standardized sets may not be equal to that reported in the source publication and this may occur for a variety of reasons. Gene identifiers reported in the article maybe have been retired, reported probes may now be recognized as non-specific or they may map to multiple genes, or gene identifiers maybe invalid due to inaccurate reporting or to MS Excel gene name conversion errors (21). GeneSigDB users have the option of seeing either the original or standardized versions of a gene set and in the current release version users can compare the original and standardized lists side-by-side. In addition, we have improved the visibility of unmapped genes so that users can identify unmapped genes in a GeneSigDB standardized table
Gene signatures in the database are identified by unique SigIDs and SigNames. The SigIDs combine the PubMed ID of the paper from which the signature was derived and the table or figure in which it was reported. For example, SigID 11823860-SuppTable2 refers to a gene signature obtained from Table 2 in the supplementary material from an article with PMID 11823860. The SigName is designed to be a more descriptive, human-readable identifier; the SigName associated with 11823860-SuppTable2 is Breast_van'tVeer02_231genes_PoorPrognosisSignature, which indicates it is a signature of poor prognosis in breast cancer, contains 231 genes and was published by van 't Veer and colleagues in 2002.
BROWSING AND SEARCHING GeneSigDB
We provide two search tools for finding signatures in the database, one based on publications and the other based on genes. The publication search tool allows users to enter one or more search terms, such as author name, article title, journal name or keywords (such as disease type), and these are then searched against the full text of articles represented in the database to find those best meeting the search criteria. In release 4.0 we added the ability to search Medical Subject Headings (MeSH) terms associated with each publication.
Users can also search for genes and their annotated properties, including gene names and synonyms, functional classifications such as GO terms, InterPro domains, KEGG or Reactome Pathways or almost any valid gene identifiers including gene symbol, Entrez gene ID, EnsEMBL gene ID, RefSeq ID or common commercial microarray probe IDs.
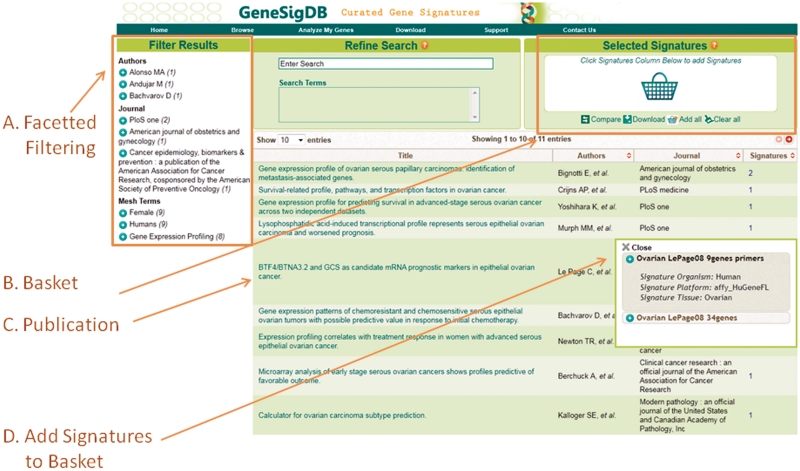
Results can be further refined by applying additional search criteria or using faceted terms associated with publication or gene search results (Figure 3). For either gene-based or publication-based searches, users can collect their results and load these into a ‘Shopping Basket’, allowing the signatures to be collected signatures using a variety of independent criteria and to then viewed, downloaded or compared (Figure 3).
Figure 3.
Screenshot showing the (A) faceting and the (B) ‘Shopping Basket’ search features when we performed a publication search for ‘serous ovarian cancer’ that returned a list of 11 publications. Further detail about (C) publications or (D) Signature can be viewed by clicking on their respective links. By default 10 results are shown but up to 100 search results can be viewed. Selecting ‘Add All’ will add the 15 gene signatures associated with the 11 displayed publications to the basket so they can be compared or downloaded.
DATA VIEWS
Clicking on a publication, gene or gene signature will open up a data type-specific view for each of these. The publication view provides information about the published article, its authors, an abstract and a list of gene signatures associated with that publication. The gene view provides annotation on a gene and a list of gene signatures which contain that gene. Each of these pages includes links to one or more gene signatures. The gene signature view now provides a dynamic table presenting both the original and standardized gene lists, each of which can be used to sort and filter the signature.
COMPARING GENE SIGNATURES
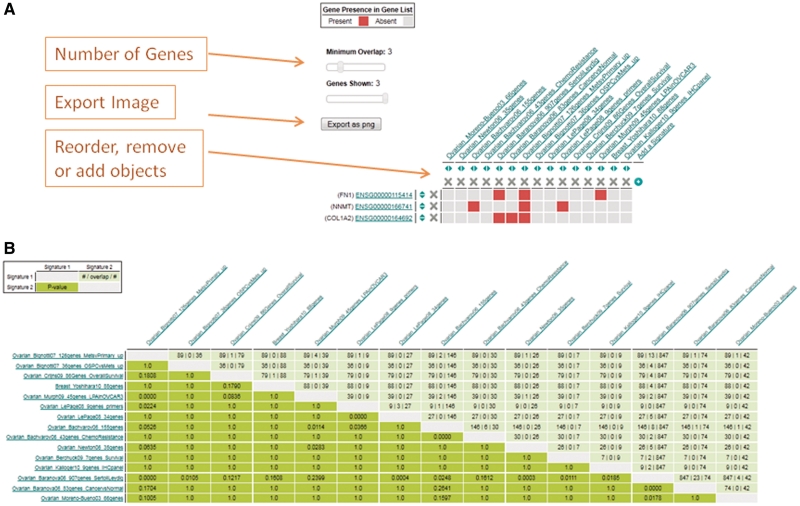
One common question is whether a particular gene set overlaps with others that have been reported. For published gene lists, we have pre-computed the pairwise similarity between all gene signatures using a one-tailed Fisher's exact test (which is equivalent to a hypergeometric distribution test) with P-values corrected for multiple testing. When a user clicks ‘Related Signatures’ from a gene signature view, or ‘Compare’ gene signatures in the Shopping Basket the most similar signatures are presented both in list and graphical form (Figure 4).
Figure 4.
Screen shots of the ‘Analyze My Gene Lists’ and ‘Comparison View’. The gene overlap between 15 serous ovarian cancer gene signatures selected in Figure 3 were analyzed. (A) shows the gene overlap between signature, where presence and absence of a gene are indicates by a red or gray pixel in the heatmap. The image can be edited, reordered or genes and gene signatures can be added or removed before the image is exported as a publication quality image. (B) shows results of a Fisher's exact test of enrichment between the 15 gene signatures. These results can also be visualized as a list.
A heatmap is used to visualize the overlaps of genes in a selection of gene signatures. In the heatmap, gray pixels indicate no overlap and red indicates two signatures sharing a common gene. Users can reorder, add or remove genes or gene signatures from the heatmap or use a sliding selector to define a minimum number of genes required for overlap. Users can also edit the heatmap column and row labels and export the heatmap as a publication quality image.
Those wishing to compare their own gene lists to the published lists in GeneSigDB can select ‘Analyze My Genes’ from the top menu bar. This brings up an interface that allows them to paste in their own gene list or to upload a file containing a gene list. If the gene list is not a list of EnsEMBL IDs, it is converted using BioMart, tested for overlap with the gene sets in the database, and reported using the table and heatmap views described above.
DOWNLOADING GeneSigDB DATA
As described above, users may perform a search to create a custom selection of gene signatures in their basket and these can be downloaded as a compressed file. In addition, all of the data in GeneSigDB is freely available for download. Users selecting ‘Download’ from the top menu bar are taken to a page where they can download the current release and previous versions of the database. The data are available in a variety of formats, including a tab-delimited flat file, GSEA gmt format and as an R/Bioconductor RData file.
PROGRAMMATIC ACCESS TO GeneSigDB DATA
Release 4.0 of GeneSigDB includes expanded programmatic access to the database through a Java RESTful web service. We use the reference implementation of JAX-RS found at Glassfish (Jersey: https://jersey.dev.java.net) to provide the REST HTTP functionality and use the Glassfish reference implementation of JAXB (JAXB: https://jaxb.dev.java.net), for the XML transformation. GeneSigDB provides REST services to retrieve each of the major objects in GeneSigDB (GeneSignature, Gene and Publication) along with all of their ancillary member objects. These objects are in either XML or JSON format. The REST request is made over HTTP by creating a URL with an embedded key that will then GET the specified resource. The Accepts portion of the HTTP request header will determine the MIME (and format) of the response. Further details and examples of how these queries should be constructed are available in GeneSigDB online documentation.
ADDING LINK OUT RESOURCES TO ADD VALUE TO GeneSigDB
In building GeneSigDB, we chose to report the published signatures rather than attempt to re-analyze the original data described in each of the manuscripts we extract from PubMed. Not only is re-analysis technically challenging because of incomplete metadata, but there are other projects that attempt to do this including Oncomine, Exalt (22) the Gene Expression Atlas (GXA) project (23) and our OncoSurf project, which is focused on finding signatures that predict survival (http://cccb.dfci.harvard.edu/oncosurf/).
The most comprehensive resource is GXA (http://www.ebi.ac.uk/gxa) whose developers have reanalyzed over 5500 gene expression and RNA-sequencing studies to identify expression profiles in 19 000 cellular or clinical phenotypes. In partnership with GXA, we are providing ‘link out’ access to GXA, providing visualization of the GeneSigDB gene sets so that users can easily see which genes within a signature are significantly associated with specific phenotypes in GXA.
SUBMITTING DATA FOR PUBLICATION IN GeneSigDB
Although we have been accepting submission to GeneSigDB by email, Release 4.0 includes a web form for signature submission. Users can also use this form to suggest updates to gene signatures currently in GeneSigDB.
USER CASES STUDIES
Although GeneSigDB is a relatively new database, it has already been used to advance our understanding of cancer and disease in new and interesting ways. Abba and colleagues used GeneSigDB 2.0 to retrieve breast cancer gene signatures (n = 42) to identify the 117 most common genes across those signatures. They found the common genes to be enriched for those associated with response to steroid hormone stimulus, and the cell cycle. Their meta-signature of the 42 GeneSigDB gene signatures was capable of predicting overall survival (P < 0.0001) and relapse-free survival (P < 0.0001) in patients with early-stage breast carcinoma. GeneSigDB has also been used to develop methods for Transcription factor binding site analysis (24) and graph theory algorithms (25)
ARCHITECTURE OF WEB INTERFACE TO GeneSigDB
The web interface to GeneSigDB (http://compbio.dfci.harvard.edu/genesigdb) is based on HTML, CSS, Javascript, JSP, XML and Java 1.6 technologies. The application runs on an Apache Tomcat 6 web application server running on a CentOS 5 Linux server. Front-end interactivity makes use of the jQuery 1.4+ Javascript library and server-side processing is based on the Apache Solr Enterprise Search Server 1.4. GeneSigDB is based on a hybrid approach between traditional database technologies and Solr indexes to catalog and organize the data. Search functions are performed using the Solr server to create a high-performance search engine. Other site functions are often performed using a database backend. Both the database and the indexes contain the same information, but they are organized in different ways to take advantage of the relative strengths of each technology.
SUMMARY AND FUTURE DIRECTIONS
GeneSigDB addresses the important need within the community to standardize gene expression signatures so they can easily be compared to each other and used in GSA. The current release represents an almost 8-fold increase in the number of gene sets from the first version and includes 3515 gene signatures derived from the analysis of cancer, stem cells, immune system function, development and lung disease; at present it is the largest source of cancer-specific gene signatures available. Based on the needs of our users, we have made considerable efforts to increase the functionality of the GeneSigDB website to facilitate mining and analysis of gene signatures.
In the future, we hope to expand GeneSigDB functionality to provide links so that users can analyze connections between genes using our predictive networks application (http://www.predictivenetworks.org) and test whether a gene signature is prognostic or associated with a known SNP using OncoSurf (http://cccb.dfci.harvard.edu/oncosurf).
Although there have been other attempts to catalog gene sets, we believe that GeneSigDB represents a significant advance in both the quantity of gene signature data we have amassed and the quality of the analysis we perform. By standardizing both the way we refer to these gene sets and the manner in which they are mapped to standard formats, we have created an infrastructure that can be scaled and extended to capture the growing number of genomic profiles that are being created and published. In a time when we as a community have become aware of the need for reproducible research, such standardization is essential to assure that the results of genomic studies can be broadly used and replicated in independent analysis. Further, the availability of standardized signatures creates an opportunity for us to more fully leverage the prior knowledge that has been gained by expert analysis of individual studies so that we can more rapidly advance our understanding of the nature of a broad range of human diseases.
FUNDING
This work was supported by National Institutes for Health National Library of Medicine (1R01 LM010129), National Cancer Institute (1U19 CA148065), Genome Research Institute (1P50 HG004233); Dana-Farber Cancer Institute Women's Cancers Program (to A.C.C); Claudia Adams Barr foundation.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank Dr Oliver Hofmann, Prof. Winston Hide and Dr Levi Waldron for their useful suggestions, which have helped us to improve GeneSigDB.
REFERENCES
- 1.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mootha VK, Lindgren CM, Eriksson K-F, Subramanian A, Sihag S, Lehar J, Puigserver P, Carlsson E, Ridderstråle M, Laurila E, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 3.Goeman JJ, Bühlmann P. Analyzing gene expression data in terms of gene sets: methodological issues. Bioinformatics. 2007;23:980–987. doi: 10.1093/bioinformatics/btm051. [DOI] [PubMed] [Google Scholar]
- 4.Liberzon A, Subramanian A, Pinchback R, Thorvaldsdóttir H, Tamayo P, Mesirov JP. Molecular signatures database (MSigDB) 3.0. Bioinformatics. 2011;27:1739–1740. doi: 10.1093/bioinformatics/btr260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bardelli A, Velculescu VE. Mutational analysis of gene families in human cancer. Curr. Opin. Genet. Dev. 2005;15:5–12. doi: 10.1016/j.gde.2004.12.009. [DOI] [PubMed] [Google Scholar]
- 6.Al-Shahrour F, Arbiza L, Dopazo H, Huerta-Cepas J, Mínguez P, Montaner D, Dopazo J. From genes to functional classes in the study of biological systems. BMC Bioinformatics. 2007;8:114. doi: 10.1186/1471-2105-8-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu C, Delano DL, Mitro N, Su SV, Janes J, McClurg P, Batalov S, Welch GL, Zhang J, Orth AP, et al. Gene set enrichment in eQTL data identifies novel annotations and pathway regulators. PLoS Genet. 2008;4:e1000070. doi: 10.1371/journal.pgen.1000070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Raychaudhuri S, Plenge RM, Rossin EJ, Ng ACY, Purcell SM, Sklar P, Scolnick EM, Xavier RJ, Altshuler D, Daly MJ. Identifying relationships among genomic disease regions: predicting genes at pathogenic SNP associations and rare deletions. PLoS Genet. 2009;5:e1000534. doi: 10.1371/journal.pgen.1000534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Montaner D, Dopazo J. Multidimensional Gene Set Analysis of Genomic Data. PLoS One. 5:e.0010348. doi: 10.1371/journal.pone.0010348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Perou CM, Børresen-Dale A-L. Systems biology and genomics of breast cancer. Cold Spring Harb. Perspect. Biol. 2011;3:t.a003293. doi: 10.1101/cshperspect.a003293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhao X, Rødland EA, Sørlie T, Naume B, Langerød A, Frigessi A, Kristensen VN, Børresen-Dale A-L, Lingjærde OC. Combining gene signatures improves prediction of breast cancer survival. PLoS ONE. 2011;6:e17845. doi: 10.1371/journal.pone.0017845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Croft D, O'Kelly G, Wu G, Haw R, Gillespie M, Matthews L, Caudy M, Garapati P, Gopinath G, Jassal B, et al. Reactome: a database of reactions, pathways and biological processes. Nucleic Acids Res. 2011;39:D691–D697. doi: 10.1093/nar/gkq1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.The Reference Genome Group of the Gene Ontology Consortium. The Gene Ontology's Reference Genome Project: a unified framework for functional annotation across species. PLoS Comput. Biol. 2009;5:e1000431. doi: 10.1371/journal.pcbi.1000431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fan C, Prat A, Parker JS, Liu Y, Carey LA, Troester MA, Perou CM. Building prognostic models for breast cancer patients using clinical variables and hundreds of gene expression signatures. BMC Med. Genomics. 2011;4:3. doi: 10.1186/1755-8794-4-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Parkinson H, Kapushesky M, Kolesnikov N, Rustici G, Shojatalab M, Abeygunawardena N, Berube H, Dylag M, Emam I, Farne A, et al. ArrayExpress update–from an archive of functional genomics experiments to the atlas of gene expression. Nucleic Acids Res. 2009;37:D868–D872. doi: 10.1093/nar/gkn889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barrett T, Troup DB, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH, Sherman PM, et al. NCBI GEO: archive for functional genomics data sets—10 years on. Nucleic Acids Res. 2010;39:D1005–D1010. doi: 10.1093/nar/gkq1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Culhane AC, Schwarzl T, Sultana R, Picard KC, Picard SC, Lu TH, Franklin KR, French SJ, Papenhausen G, Correll M, et al. GeneSigDB–a curated database of gene expression signatures. Nucleic Acids Res. 2010;38:D716–D725. doi: 10.1093/nar/gkp1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liberzon A, Subramanian A, Pinchback R, Thorvaldsdóttir H, Tamayo P, Mesirov JP. Molecular signatures database (MSigDB) 3.0. Bioinformatics. 2011;27:1739–1740. doi: 10.1093/bioinformatics/btr260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dietmann S, Lee W, Wong P, Rodchenkov I, Antonov AV. CCancer: a bird's eye view on gene lists reported in cancer-related studies. Nucleic Acids Res. 2010;38:W118–W123. doi: 10.1093/nar/gkq515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haider S, Ballester B, Smedley D, Zhang J, Rice P, Kasprzyk A. BioMart Central Portal—unified access to biological data. Nucleic Acids Res. 2009;37:W23–W27. doi: 10.1093/nar/gkp265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zeeberg BR, Riss J, Kane DW, Bussey KJ, Uchio E, Linehan WM, Barrett JC, Weinstein JN. Mistaken identifiers: gene name errors can be introduced inadvertently when using Excel in bioinformatics. BMC Bioinformatics. 2004;5:80. doi: 10.1186/1471-2105-5-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu J, Qiu Q, Xie L, Fullerton J, Yu J, Shyr Y, George AL, Yi Y. Web-based interrogation of gene expression signatures using EXALT. BMC Bioinformatics. 2009;10:420. doi: 10.1186/1471-2105-10-420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kapushesky M, Emam I, Holloway E, Kurnosov P, Zorin A, Malone J, Rustici G, Williams E, Parkinson H, Brazma A. Gene expression atlas at the European bioinformatics institute. Nucleic Acids Res. 2010;38:D690–D698. doi: 10.1093/nar/gkp936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Van de Sande B, Atak ZK, Aerts S. Discovery of regulators for co-expressed human genes using large sequence search spaces. Bioinformatics for Regulatory Genomics (BioRegSIG) Special Interest Group (SIG) at ISMB. 2011 http://light.ece.ohio.edu/bioreg/2011/pages/vandesande.pdf. (16 July 2011, date last accessed) [Google Scholar]
- 25.Georgii E, Tsuda K, Schölkopf B. Multi-way set enumeration in weight tensors. Machine Learning. 2011;82:123–155. [Google Scholar]