Abstract
Large numbers of mass spectrometry proteomics studies are being conducted to understand all types of biological processes. The size and complexity of proteomics data hinders efforts to easily share, integrate, query and compare the studies. The Model Organism Protein Expression Database (MOPED, htttp://moped.proteinspire.org) is a new and expanding proteomics resource that enables rapid browsing of protein expression information from publicly available studies on humans and model organisms. MOPED is designed to simplify the comparison and sharing of proteomics data for the greater research community. MOPED uniquely provides protein level expression data, meta-analysis capabilities and quantitative data from standardized analysis. Data can be queried for specific proteins, browsed based on organism, tissue, localization and condition and sorted by false discovery rate and expression. MOPED empowers users to visualize their own expression data and compare it with existing studies. Further, MOPED links to various protein and pathway databases, including GeneCards, Entrez, UniProt, KEGG and Reactome. The current version of MOPED contains over 43 000 proteins with at least one spectral match and more than 11 million high certainty spectra.
INTRODUCTION
Protein expression, the presence or quantity of a protein in a biological sample, is one of the key measures essential for understanding biological processes. The data serve as a snapshot of the state of an organism at the time of sample collection. Notably, aberrant protein expression patterns in disease states may be indicative of the mis-regulations associated with the disease. MOPED (http://moped.proteinspire.org) was motivated, in part, by the idea that easy public access to protein expression data will enable scientists to better identify and understand protein expression patterns that are related to significant diseases and biological processes.
Mass spectrometry-based proteomics is the most common approach used to survey complex samples for the presence of proteins and their expression (1,2). To provide ample context for the data contained in MOPED, we briefly describe a proteomics workflow.
Prior to analysis by mass spectrometry, proteins are typically digested into their peptide components. Search engines such as Sequest, Mascot, X!Tandem and OMSSA match the spectra generated by tandem mass spectrometry with peptides from a target protein sequence database (3–6). Due to the highly complex nature of protein samples and their processing, as well as mass spectrometry instrumentation, approaches and analysis, peptide spectral matches are associated with varying degrees of uncertainty (7–9). Once peptide spectral matches are formed, the peptides are amalgamated into protein identifications with associated measures of statistical certainty. Commonly, peptide spectral matches are performed against decoy databases generated by reversing or randomizing the target database to estimate the false discovery rate (FDR) associated with protein and peptide identifications (10,11).
From these searches, estimates of protein expression can be determined by using measures such as spectra counts (the number of identified spectra which correspond to a specific protein), sequence coverage and peak areas or intensities (12,13). Expression in mass spectrometry proteomics experiments can be measured dichotomously in terms of the certainty of a protein being present or with quantitative measures that reflect the protein's concentration. Relative expression measures are used for comparing the relative amounts of the same protein across different conditions. Absolute expression, the quantification of different proteins within the same sample is difficult to measure in part due to variability in individual protein responses to mass spectrometry assay methods.
A number of websites provide host services for massive proteomics datasets (14–17). Although these repositories are excellent resources for accessing raw data and quick experimental summaries, they neither provide protein expression data, nor do they allow for a standardized comparison of expression levels across tissues, localizations and conditions. Furthermore, the extreme scale of data in these repositories makes meta-analysis and even simple querying of these datasets a staggering challenge, often worthy of its own publication (18,19). Such meta-analysis typically requires the download of raw data, whose volume is often measured in terabytes, and analysis of these data through a computationally intensive proteomics workflow. In cases where summary information is available, these data may be in varying formats, have been processed through non-standard pipelines and often provide limited or non-comparable statistical measures of protein identification certainty. Additionally, proteome profiles from other resources omit the relevant expression information (20).
The aforementioned challenges hinder the utilization of publicly available proteomics data. Enabling researchers to access these data in an effective manner is an important challenge in proteomics. MOPED complements the availability of raw data from other resources by presenting standardized data analysis and enabling the user to view experimental data relative to existing expression profiles across many different tissues, localizations and conditions (21).
Where there are multiple experimental datasets for a given combination of organism, tissue, localization and condition, a meta-analysis is provided based on the recently published approach (18). The simple format of the MOPED data and the straightforward approach to meta-analysis allows for the uncomplicated combination of proteomics datasets. These features and comparisons empower the user to make meaningful statements about identified proteins with respect to the existing knowledge-base.
DATABASE CONTENT
Expression data
The core component of MOPED's database is the repository of expression information from public proteomics datasets. By storing and displaying essential summary information without requiring the user to download any files, MOPED simplifies access to the proteomics data. To maintain statistical integrity, MOPED requires that statistical measures be provided for each protein identification, including the protein FDR and spectral counts. A full list of required measures is found in Table 1. Users may submit data to MOPED by providing either raw files or pre-processed data. Currently, all data displayed in MOPED were analyzed using the standardized data analysis and statistical methods of the SPIRE pipeline (21,22).
Table 1.
The fields required for each protein expression data point in MOPED
| Statistic | Definition |
|---|---|
| Expression percentile | The percentile (0–100%) corresponding to the protein expression level in this experiment |
| Normalized expression | Number of spectra counts divided by sequence length normalized to the maximum expression value in the experiment (0–1) |
| FDR | Cumulative FDR threshold for protein identification |
| Spectral count | The number of unique spectra identified which correspond to the identified proteins. |
| Unique peptides | Number of unique peptide sequences identified |
| Sequence coverage | Percentage of the protein sequence covered by identified peptide sequences |
Meta-data
A major problem when accessing public data is a lack of specificity from data providers about experimental protocols. To prevent this frustration, MOPED requires a minimum amount of meta-data that must be included with each dataset. At the experiment level, users must supply a brief experimental description, the source organism from the NCBI taxon database and any applicable journal references (23). Additionally, each protein identification is associated with a tissue, localization and condition which align with the BRENDA Tissue Ontology, Cell Type Ontology and Disease Ontology, respectively (24–26).
Organisms
MOPED contains information on both humans and model organisms. Not only does studying model organisms increase our understanding of biological systems, but also studies of model organisms can inform our knowledge of homologous systems in humans and other species (27). Thus far, MOPED contains data from four of the most studied organisms: Homo sapiens (human), Mus musculus (mouse), Caenorhabditis elegans (worm) and Saccharomyces cerevisiae (yeast).
Protein information
To maximize information content, MOPED has been built to link out to many of the most popular and useful data resources. In terms of protein identifiers, MOPED has universal links to the heavily utilized UniProt and NCBI databases and organism-specific links to the authoritative WormBase and Saccharomyces Genome Database (28–31). A symbiotic relationship has been established whereby, MOPED links to GeneCards and GeneCards displays MOPED’s data (32). MOPED contains an innovative database that extends coverage of proteins to pathway databases (KEGG, Reactome, Metacyc, PANTHER and SEED) using orthologous groups of proteins specified by both the aforementioned pathways databases and eggNOG (33–38). In total, MOPED links to 10 external databases.
Release statistics
As of 10 November 2011, MOPED contains 43 794 proteins with at least one high certainty spectral match, 23 167 proteins with an FDR<1% and more than 11 million spectra (39). These data come from 35 experiments on 4 organisms covering 13 tissues, 21 localizations and 10 conditions. Organism-specific release statistics are in Table 2. In addition to individual experiments, the database also contains meta-analyses of yeast and worm data based upon the recently published approach to meta-analysis (18).
Table 2.
Release statistics as of 10 November 2011
| Species | Proteins with at least one spectral match | Proteins with <1% FDR | High confidence spectra |
|---|---|---|---|
| Homo sapiens (human) | 15 847 | 6102 | 3 906 048 |
| Mus musculus (mouse) | 10 308 | 5935 | 2 650 237 |
| Caenorhabditis elegans (worm) | 10 922 | 7383 | 1 979 744 |
| Saccharomyces cerevisiae (yeast) | 6717 | 3747 | 2 809 390 |
| Total | 43 794 | 23 167 | 11 345 419 |
USER INTERFACE
MOPED front page
The MOPED front page (http://moped.proteinspire.org) provides a description of the MOPED resource and contains tabs to access database search, upload data and view help files.
MOPED search view
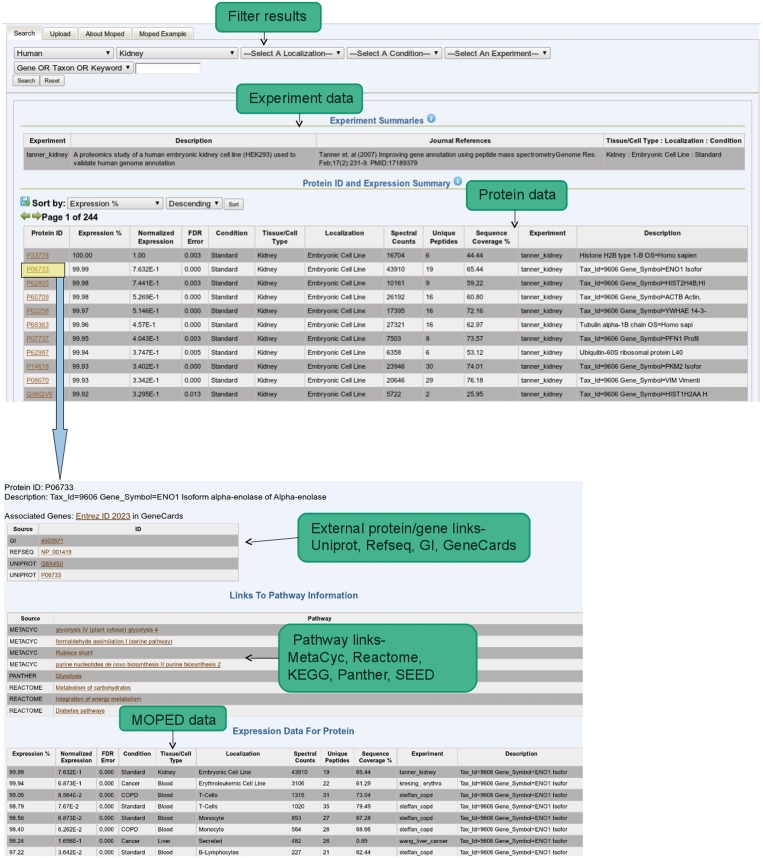
MOPED's access point to proteomics data is located in the ‘Search’ tab. From this view, users are able to access the entirety of MOPED's expression database (Figure 1, top). Protein expression data can be both browsed by categories such as organism, tissue and localization and queried by protein ID and keywords. After the user has selected filters, clicking the ‘Search’ button quickly renders all matching expression data points and associated meta-data. Most of the search view is dominated by the ‘Protein ID and Expression Summary’ section which displays expression data resulting from the user's query. Each row in the expression summary table displays all statistical information contained in Table 1, as well as experimental meta-data. Complete protein annotations can be viewed by hovering over either the protein IDs or partial annotations. The set of meta-data corresponding to all displayed expression information is summarized under the separate ‘Experiment Summaries’ table. The filtering capabilities at the top of the MOPED interface's Search tab allows users to query on these different experiments.
Figure 1.
MOPED views. The main MOPED view, on top and the protein view, on bottom. Clicking on links for an identified protein in the main MOPED view brings up the protein view. In this example, P06733 has been selected from the main MOPED view.
MOPED protein view
Clicking on a protein ID from any tab allows the user to open a page containing all stored information related to that protein, including the protein annotation, links to protein and pathway databases and identifications of that protein in other MOPED experiments (Figure 1, bottom).
The primary advantage of MOPED's protein view over other databases is the presentation of expression data from many experiments side by side. On the protein page, MOPED automatically displays the expression information for that protein in every single experiment contained in MOPED (Figure 1, bottom). Ideally, this information will enable the user to identify meaningful expression patterns across different conditions. The same expression information has been incorporated with both GeneCards (human data only) and SPIRE (32,21).
MOPED upload
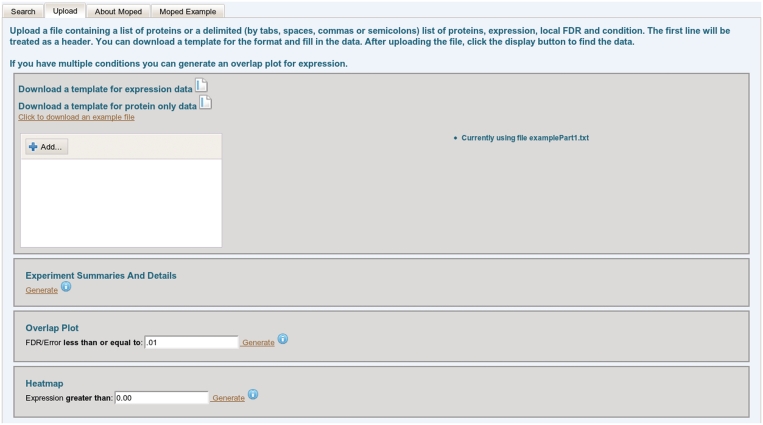
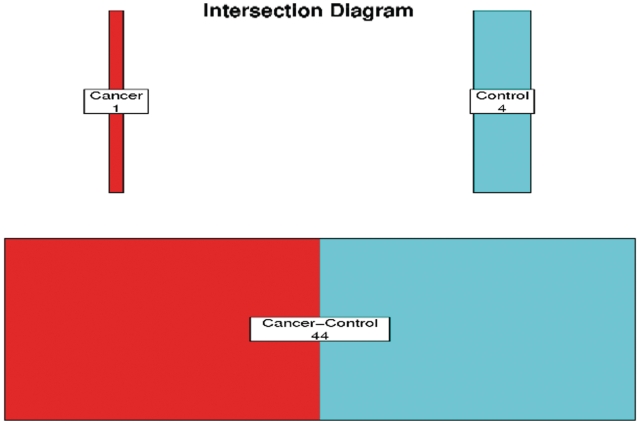
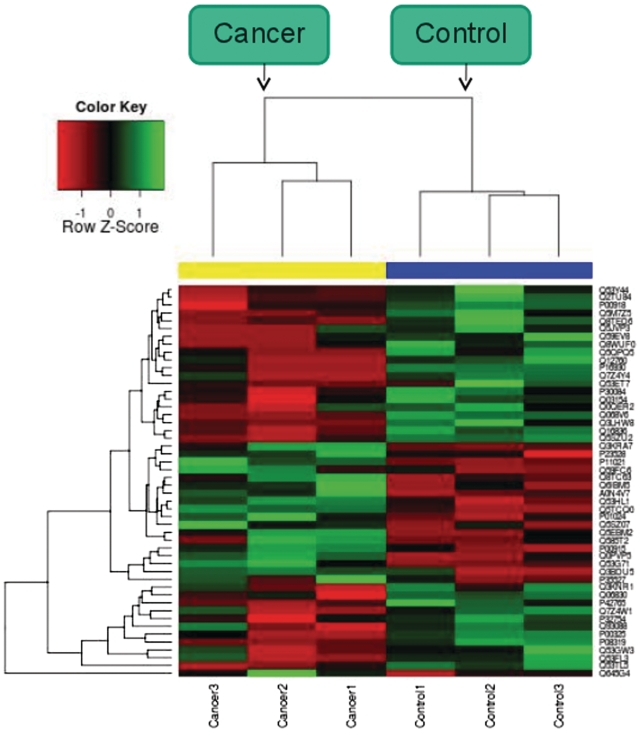
Through the upload tab, users can compare their experimental data with the data contained in the MOPED servers. User upload of data automatically filters MOPED data to display only those proteins which were identified in the user's experiment. For identification only queries, users are able to upload a list of UniProt protein identifiers. For expression based queries, users may upload UniProt protein identifiers, expression and FDR values and condition names. Once this information has been uploaded, the user can experiment with several functionalities in the Upload tab (Figure 2). MOPED displays the data for proteins identified in both the user's experiment and experiments in the MOPED servers. These data may be interrogated in the same manner as the MOPED search page. For identification visualization, MOPED separates user data based on condition and generates overlap plots of the identifications with dynamic thresholding by protein FDR (Figure 3). For expression visualization, MOPED dynamically generates heatmaps of the user-uploaded data with user-specified expression value thresholding (Figure 4).
Figure 2.
Upload tab. Users may upload their own data through the upload tab. These data can then be visualized by clicking any of the ‘Generate’ links under their associated functionalities. Experiment summaries and details create a view at the bottom of the screen akin to the view in Figure 1. The overlap plot and heatmap views are seen in Figure 3 and Figure 4, respectively.
Figure 3.
Overlap plot. An overlap plot generated for data from Ref. (42) with two conditions, cancer and control.
Figure 4.
Overlap plot. An overlap plot generated for data from Ref. (42) with two conditions, cancer and control.
MOPED documentation
MOPED provides a comprehensive help file and a tutorial example to clarify the usage and highlight its features. This documentation is accessible under the Help tab and comes in the form of two pdf files. The tutorial contains real data examples.
FUTURE DIRECTIONS
Increased data and public data submission
MOPED is currently involved in a number of collaborations that will dramatically increase the amount of proteomics data available. Though all MOPED data are currently loaded in-house, work is in progress to create an interface for public submission of proteomics expression data. Users will be able to fulfill publication and grant requirements for data preservation by uploading their datasets to MOPED. Researchers interested in submitting their data are invited to contact the MOPED team at moped@proteinspire.org. In addition to increasing the number of protein identification experiments, MOPED plans to utilize data from relative expression experiments, providing users with expression ratios and statistical significance for many different condition comparisons.
Increased visualization
MOPED remains under continuous development to improve all components of the user experience. Currently, work is underway to develop a plug-in for Cytoscape that provides pathway level visualization of the experimental data currently residing in MOPED (40). The goal is to maximize the user's knowledge of fluctuating patterns of pathway regulation (Supplementary Figure S5). Additionally, scripts are being developed to dynamically visualize experimental expression relative to the MOPED experiments (Supplementary Figure S6).
Integration of other omics data
While proteomics data provides comprehensive insight into cellular mechanisms at the protein level, combining proteomics knowledge with other omics disciplines stands to develop a more complete understanding of complex biological systems. Metabolomics, transcriptomics, lipidomics and genomics are notable disciplines for which integrated analysis with proteomics is a natural extension. For example, proteomics data from MOPED could be linked with transcriptomics data from GEO for common organ, tissue, localization and condition combinations (41).
DISCUSSION
Currently, proteomics datasets are either scattered throughout individual data repositories or trapped within labs’ own databases. Knowledge discovery is often obscured by bulky datasets, non-standard formats, missing meta-data and limited access to data. MOPED presents a solution which addresses these challenges. MOPED provides essential statistical summaries and a number of query and visualization tools to relate the findings to those observed in other experiments. Patterns of expression within and across sample sets can be visualized, proteins of interest can be directly queried and condition-specific expression data can be browsed. As community resource, MOPED will increase reliable data proliferation and make analysis more comprehensive.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures 5 and 6.
FUNDING
National Science Foundation (DBI grant 0544757); National Institutes of Health (NIGMS grant 5R01 GM076680-02, NIDDK grants UO1 DK072473, 1U01DK089571-01); the McMillen Foundation (grant to E.K.). Funding for open access charge: Seattle Children's.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We would like to thank Elizabeth Stewart, Chris Howard, Chris Moss, Courtney MacNealy-Koch and Carey Sheu for their comments, critical assessment and help in developing MOPED and this manuscript. Marilyn Safran, Gil Stelzer, the GeneCards team, Biaoyang Lin, Eric Deutsch, Ruedi Aebersold, Matthias Mann, Jurgen Cox, Gordon Anderson, Tom Metz and Richard Smith for their assistance in gathering data and input into the development of MOPED and this manuscript. We also thank the Executive Editor and the Referees for their insightful comments that helped improve the quality of this manuscript.
REFERENCES
- 1.Aebersold R, Mann M. Mass spectrometry-based proteomics. Nature. 2003;422:198–207. doi: 10.1038/nature01511. [DOI] [PubMed] [Google Scholar]
- 2.Griffin TJ, Goodlett DR, Aebersold R. Advances in proteome analysis by mass spectrometry. Curr. Opin. Biotechnol. 2001;12:607–612. doi: 10.1016/s0958-1669(01)00268-3. [DOI] [PubMed] [Google Scholar]
- 3.Eng JK, McCormack AL, Yates JR., III An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass Spectr. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 4.Perkins DN, Pappin DJ, Creasy DM, Cottrell JS. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999;20:3551–3567. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 5.Fenyö D, Beavis RC. A method for assessing the statistical significance of mass spectrometry-based protein identifications using general scoring schemes. Anal. Chem. 2003;75:768–774. doi: 10.1021/ac0258709. [DOI] [PubMed] [Google Scholar]
- 6.Geer LY, Markey SP, Kowalak JA, Wagner L, Xu M, Maynard DM, Yang X, Shi W, Bryant SH. Open mass spectrometry search algorithm. J. Proteome Res. 2004;3:958–964. doi: 10.1021/pr0499491. [DOI] [PubMed] [Google Scholar]
- 7.Keller A, Nesvizhskii AI, Kolker E, Aebersold R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal. Chem. 2002;74:5383–5392. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- 8.Kolker E, Higdon R, Hogan JM. Protein identification and expression analysis using mass spectrometry. Trends Microbiol. 2006;14:229–235. doi: 10.1016/j.tim.2006.03.005. [DOI] [PubMed] [Google Scholar]
- 9.Nesvizhskii AI, Keller A, Kolker E, Aebersold R. A statistical model for identifying proteins by tandem mass spectrometry. Anal. Chem. 2003;75:4646–4658. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- 10.Elias JE, Gygi SP. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat. Methods. 2007;4:207–214. doi: 10.1038/nmeth1019. [DOI] [PubMed] [Google Scholar]
- 11.Higdon R, Hogan JM, Van Belle G, Kolker E. Randomized sequence databases for tandem mass spectrometry peptide and protein identification. OMICS. 2005;9:364–379. doi: 10.1089/omi.2005.9.364. [DOI] [PubMed] [Google Scholar]
- 12.Bantscheff M, Schirle M, Sweetman G, Rick J, Kuster B. Quantitative mass spectrometry in proteomics: a critical review. Anal. Bioanal. Chem. 2007;389:1017–1031. doi: 10.1007/s00216-007-1486-6. [DOI] [PubMed] [Google Scholar]
- 13.Liu H, Sadygov RG, Yates JR. A Model for random sampling and estimation of relative protein abundance in shotgun proteomics. Anal. Chem. 2004;76:4193–4201. doi: 10.1021/ac0498563. [DOI] [PubMed] [Google Scholar]
- 14.Deutsch EW, Lam H, Aebersold R. PeptideAtlas: a resource for target selection for emerging targeted proteomics workflows. EMBO Rep. 2008;9:429–434. doi: 10.1038/embor.2008.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vizcaíno JA, Côté R, Reisinger F, Foster JM, Mueller M, Rameseder J, Hermjakob H, Martens L. A guide to the Proteomics Identifications Database proteomics data repository. Proteomics. 2009;9:4276–4283. doi: 10.1002/pmic.200900402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Craig R, Cortens JP, Beavis RC. Open source system for analyzing, validating, and storing protein identification data. J. Proteome Res. 2004;3:1234–1242. doi: 10.1021/pr049882h. [DOI] [PubMed] [Google Scholar]
- 17.Hill JA, Smith BE, Papoulias PG, Andrews PC. ProteomeCommons.org collaborative annotation and project management resource integrated with the tranch repository. J. Proteome Res. 2010;9:2809–2811. doi: 10.1021/pr1000972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Higdon R, Haynes W, Kolker E. Meta-analysis for protein identification: a case study on yeast data. OMICS. 2010;14:309–314. doi: 10.1089/omi.2010.0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang Q, Faca V, Hanash S. Mining the plasma proteome for disease applications across seven logs of protein abundance. J. Proteome Res. 2011;10:46–50. doi: 10.1021/pr101052y. [DOI] [PubMed] [Google Scholar]
- 20.Gnad F, Oroshi M, Birney E, Mann M. MAPU 2.0: high-accuracy proteomes mapped to genomes. Nucleic Acids Res. 2009;39:D902–D906. doi: 10.1093/nar/gkn773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kolker E, Higdon R, Welch D, Bauman A, Stewart E, Haynes W, Broomall W, Kolker N. SPIRE: Systematic protein investigative research environment. J. Proteomics. 2011 doi: 10.1016/j.jprot.2011.05.009. May 13 (doi: 10.1016/j.jprot.2011.05.009; epub ahead of print) [DOI] [PubMed] [Google Scholar]
- 22.Higdon R, Reiter L, Hather G, Haynes W, Kolker N, Stewart E, Bauman AT, Picotti P, Schmidt A, van Belle G, et al. IPM: An integrated protein model for false discovery rate estimation and identification in high-throughput proteomics. J. Proteomics. 2011 doi: 10.1016/j.jprot.2011.06.003. doi: 10.1016/j.jprot.2011.06.003. [DOI] [PubMed] [Google Scholar]
- 23.Sayers EW, Barrett T, Benson DA, Bolton E, Bryant SH, Canese K, Chetvernin V, Church DM, DiCuccio M, Federhen S, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2011;39:D38–D51. doi: 10.1093/nar/gkq1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gremse M, Chang A, Schomburg I, Grote A, Scheer M, Ebeling C, Schomburg D. The BRENDA Tissue Ontology (BTO): the first all-integrating ontology of all organisms for enzyme sources. Nucleic Acids Res. 2011;39:D507–D513. doi: 10.1093/nar/gkq968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bard J, Rhee SY, Ashburner M. An ontology for cell types. Genome Biol. 2005;6:R21. doi: 10.1186/gb-2005-6-2-r21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Osborne JD, Flatow J, Holko M, Lin SM, Kibbe WA, Zhu L, Danila MI, Feng G, Chisholm RL. Annotating the human genome with Disease Ontology. BMC Genomics. 2009;10:S6. doi: 10.1186/1471-2164-10-S1-S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Botstein D. Genetics: yeast as a model organism. Science. 1997;277:1259–1260. doi: 10.1126/science.277.5330.1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pruitt KD, Tatusova T, Klimke W, Maglott DR. NCBI Reference Sequences: current status, policy and new initiatives. Nucleic Acids Res. 2009;37:D32–D36. doi: 10.1093/nar/gkn721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Uniprot Consortium. Ongoing and future developments at the Universal Protein Resource. Nucleic Acids Res. 2011;39:D214–D219. doi: 10.1093/nar/gkq1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Engel SR, Balakrishnan R, Binkley G, Christie KR, Costanzo MC, Dwight SS, Fisk DG, Hirschman JE, Hitz BC, Hong EL, et al. Saccharomyces Genome Database provides mutant phenotype data. Nucleic Acids Res. 2010;38:D433–D436. doi: 10.1093/nar/gkp917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Harris TW, Antoshechkin I, Bieri T, Blasiar D, Chan J, Chen WJ, De La Cruz N, Davis P, Duesbury M, Fang R, et al. WormBase: a comprehensive resource for nematode research. Nucleic Acids Res. 2010;38:D463–D467. doi: 10.1093/nar/gkp952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Safran M, Dalah I, Alexander J, Rosen N, Iny Stein T, Shmoish M, Nativ N, Bahir I, Doniger T, Krug H, et al. GeneCards Version 3: the human gene integrator. Database. 2010;2010 doi: 10.1093/database/baq020. doi: 10.1093/database/baq020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kanehisa M, Goto S, Furumichi M, Tanabe M, Hirakawa M. KEGG for representation and analysis of molecular networks involving diseases and drugs. Nucleic Acids Res. 2010;38:D355–D360. doi: 10.1093/nar/gkp896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Croft D, O'Kelly G, Wu G, Haw R, Gillespie M, Matthews L, Caudy M, Garapati P, Gopinath G, Jassal B, et al. Reactome: a database of reactions, pathways and biological processes. Nucleic Acids Res. 2011;39:D691–D697. doi: 10.1093/nar/gkq1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Caspi R, Altman T, Dale JM, Dreher K, Fulcher CA, Gilham F, Kaipa P, Karthikeyan AS, Kothari A, Krummenacker M, et al. The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases. Nucleic Acids Res. 2010;38:D473–D479. doi: 10.1093/nar/gkp875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mi H, Dong Q, Muruganujan A, Gaudet P, Lewis S, Thomas PD. PANTHER version 7: improved phylogenetic trees, orthologs and collaboration with the Gene Ontology Consortium. Nucleic Acids Res. 2009;38:D204–D210. doi: 10.1093/nar/gkp1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Overbeek R, Begley T, Butler RM, Choudhuri JV, Chuang H-Y, Cohoon M, de Crécy-Lagard V, Diaz N, Disz T, Edwards R, et al. The subsystems approach to genome annotation and its use in the project to annotate 1000 genomes. Nucleic Acids Res. 2005;33:5691–5702. doi: 10.1093/nar/gki866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Muller J, Szklarczyk D, Julien P, Letunic I, Roth A, Kuhn M, Powell S, von Mering C, Doerks T, Jensen LJ, et al. eggNOG v2.0: extending the evolutionary genealogy of genes with enhanced non-supervised orthologous groups, species and functional annotations. Nucleic Acids Res. 2010;38: D190–D195. doi: 10.1093/nar/gkp951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hather G, Higdon R, Bauman A, von Haller PD, Kolker E. Estimating false discovery rates for peptide and protein identifications using randomized databases. Proteomics. 2010;10:2369–2376. doi: 10.1002/pmic.200900619. [DOI] [PubMed] [Google Scholar]
- 40.Smoot ME, Ono K, Ruscheinski J, Wang P-L, Ideker T. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics. 2011;27:431–432. doi: 10.1093/bioinformatics/btq675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Barrett T, Troup DB, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH, Sherman PM, et al. NCBI GEO: archive for functional genomics data sets–10 years on. Nucleic Acids Res. 2011;39:D1005–D1010. doi: 10.1093/nar/gkq1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang J, Gao F, Mo F, Hong X, Wang H, Zheng S, Lin B. Identification of CHI3L1 and MASP2 as a biomarker pair for liver cancer through integrative secretome and transcriptome analysis. Proteom.– Clin. Appl. 2009;3:541–551. [Google Scholar]