Abstract
Close to 60% of protein sequences tracked in comprehensive databases can be mapped to a known three-dimensional (3D) structure by standard sequence similarity searches. Potentially, a great deal can be learned about proteins or protein families of interest from considering 3D structure, and to this day 3D structure data may remain an underutilized resource. Here we present enhancements in the Molecular Modeling Database (MMDB) and its data presentation, specifically pertaining to biologically relevant complexes and molecular interactions. MMDB is tightly integrated with NCBI's Entrez search and retrieval system, and mirrors the contents of the Protein Data Bank. It links protein 3D structure data with sequence data, sequence classification resources and PubChem, a repository of small-molecule chemical structures and their biological activities, facilitating access to 3D structure data not only for structural biologists, but also for molecular biologists and chemists. MMDB provides a complete set of detailed and pre-computed structural alignments obtained with the VAST algorithm, and provides visualization tools for 3D structure and structure/sequence alignment via the molecular graphics viewer Cn3D. MMDB can be accessed at http://www.ncbi.nlm.nih.gov/structure.
INTRODUCTION
The three-dimensional (3D) structures of macromolecules, as collected and provided by the Protein Data Bank (PDB) (1), offer tremendous insights into molecular function at the atomic level, and often they provide direct evidence for aspects of that function by exemplifying molecular interactions between individual macromolecules, or between macromolecules and small molecules. The examination of 3D structure often provides explanations for patterns of sequence conservation observed in protein families, and 3D structure alignment can serve as a guide for constructing accurate multiple sequence alignments such as used in phylogenetic analysis.
Recently the PDB has begun to provide comprehensive data regarding the biologically relevant assemblies/complexes or quaternary structures of the macromolecules described in its records (1). In X-ray crystallography, the results of the experiment as reported to the PDB may not always describe the biologically active or relevant complex. What is submitted to the PDB archive and redistributed from the archive may contain more than one such complex, or it may contain just a fraction of the biologically relevant assembly/complex. Typically, the relevant biological assemblies/complexes are assigned by the structure authors, but they may also be provided as assigned by automated algorithms such as PISA (2). In such cases where the actual quaternary state of a molecular complex is known, the displays should feature that view rather than a view of the structure's raw data, as most database users may be more interested in biological function than in details about the data collection. Accordingly, the NCBI Molecular Modeling Database [MMDB; (3)] has been enhanced to emphasize the functional molecular complex (i.e. quaternary structure) and the interactions between its molecular components.
The IBIS (4) resource, also developed at NCBI, organizes, analyzes and predicts interaction partners and locations of binding sites in proteins. It clusters interactions that appear conserved in molecular evolution using 3D structure alignments, and ranks the clusters so as to emphasize the biologically relevant interactions. IBIS allows inference of interactions and binding sites for arbitrary proteins by sequence similarity to 3D structures in the database. The interactions in MMDB are those which are actually observed in the individual 3D structures, and constitute the underlying data for the molecular interactions inferred by IBIS.
MMDB is updated once a week, in synchrony with the PDB. The update and computation of new and additional structure neighbors usually takes no more than 2 or 3 days to complete, depending on the size of the update and the nature of the newly added structures. Currently, MMDB holds 75 800 entries, more than half of which represent molecular complexes with two or more macromolecules. Here we describe recent additions to the data tracked by MMDB and the revised structure summary pages provided by the MMDB web service.
BIOLOGICAL UNITS AND INTERACTION SCHEMATICS
The revised structure summary presentation is shown in Figure 1. MMDB now tracks biological assemblies/complexes—or ‘biological units’, as they are called on the structure summary pages—and will by default display the first or default biological unit listed in the source data. A navigation aid in the top section of the summary pages, right below the primary citation, offers three different views of the record: (i) the default biological unit; (ii) a list of all biological units, including the default, which may represent multiple copies of the biological assembly that were present in the raw data, and/or various interpretations of the biological assembly; and (iii) the asymmetric unit as the set of data submitted to the Protein Data Bank by the depositor. As the views are toggled, the detailed presentation on the page may differ, as may the options available for data download and visualization.
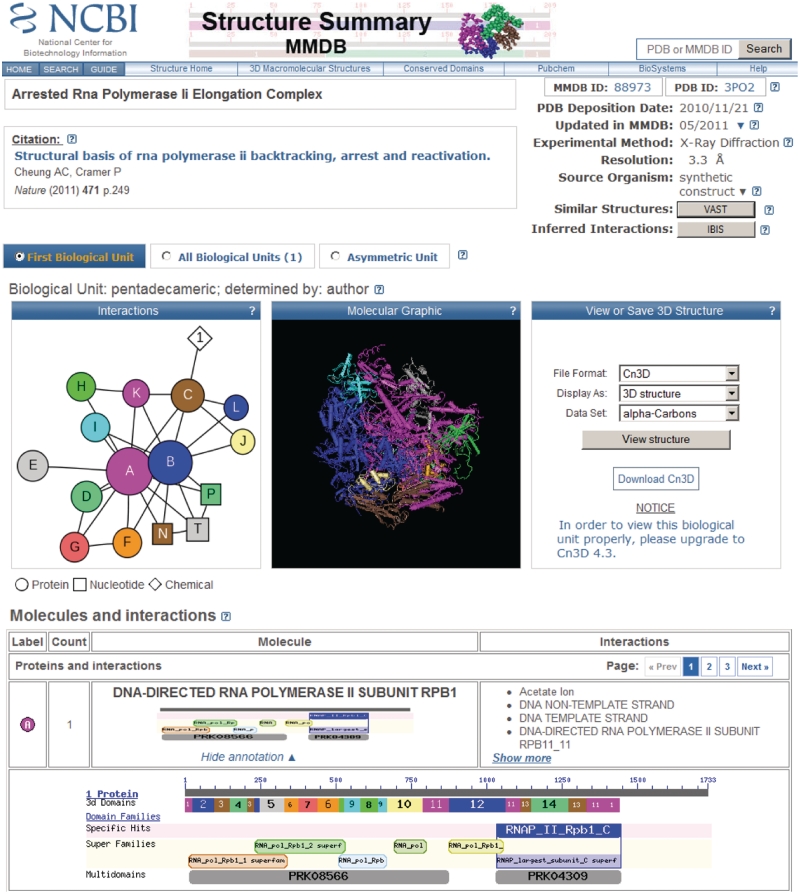
Figure 1.
Structure summary page for the MMDB entry 88973 with PDB accession 3PO2. This is a structure of yeast RNA Polymerase II complex trapped in the elongation process, and contains polypeptides, nucleic acids and ions. The table of molecules and interactions is truncated in this screenshot, with the sequence annotation view for the first polypeptide in the list expanded to full detail. The colors used to depict 3d Domains in the sequence annotation graphic are independent of the colors used to depict molecules in the interactions schematic. The complete table that is displayed on live structure summary pages provides similar information for each molecule in the structure. The page provides links to the source database (PDB), taxonomy, PubMed, Entrez/Protein, Entrez/Nucleotide, PubChem, IBIS, VAST structure neighbors and the Conserved Domain Database. The interaction schematic shows that only the two largest protein subunits of the complex interact with the nucleic acids (which represent the double-stranded DNA template and the RNA product), and that the smaller protein subunits surround the core of the complex and may interact with both or only one of the two large subunits.
3D structures of biological units may be visualized using the 3D viewer Cn3D (5), which has recently been released as a new version v4.3, to support visualization of biological units with macromolecules generated via symmetry operations. Cn3D v4.3 also comes with a wider range of features, such as side-by-side stereo, and is distributed as a helper application for the web-browser (see Table 1 below for the download URL).
Table 1.
URLs and other resources associated with MMDB
| MMDB | Database home page | http://www.ncbi.nlm.nih.gov/structure |
| MMDB FTP | Data distribution | ftp://ftp.ncbi.nih.gov/mmdb/ |
| VAST search | Find similar 3D structures via structure comparison | http://www.ncbi.nlm.nih.gov/Structure/VAST/vastsearch.html |
| IBIS | Inferred interactions | http://www.ncbi.nlm.nih.gov/Structure/ibis/ibis.cgi |
| CDD | Conserved Domains | http://www.ncbi.nlm.nih.gov/Structure/cdd/cdd.shtml |
| Cn3D | Molecular graphics viewer | http://www.ncbi.nlm.nih.gov/Structure/CN3D/cn3d.shtml |
| CBLAST | Find-related structures via sequence comparison | http://www.ncbi.nlm.nih.gov/Structure/cblast/cblast.cgi |
3D structures may also be visualized using any viewer that works with PDB file format, such as RasMol (6) and its derivatives. If the record view is set to ‘Asymmetric Unit’, the user may select between NCBI's variant of the PDB file formatted data and the original record as obtained from the PDB archive. It is now also possible to save the data for any given biological unit as a PDB formatted file, including biological units that were reconstructed by applying transformations according to crystallographic symmetry.
The structure summary pages provide a molecular graphics thumbnail which reflects the view of the biological unit (or asymmetric unit) that Cn3D will show by default if launched from the page. The default coloring has been changed from ‘secondary structure’ to ‘color by molecule’, as the presentation now emphasizes the make-up of multi-molecular complexes. However, it may be very difficult to inspect a molecular graphics snapshot—and even a live 3D visualization session—and understand whether, and to what extent, macromolecules and small molecules interact with each other in a larger, non-trivial multi-molecular assembly. To this end, MMDB now pre-computes and stores molecular interactions as derived from the imported structure data. Two biopolymers are said to be in contact, if atoms from five or more of their constituent residues are involved in close contacts (<4 Å) with atoms from the other molecule, and a similar threshold is employed to compute and track interactions between biopolymers and small molecules/chemicals. The thresholds employed are compatible with those used in the IBIS (2) resource.
Structure summary pages now also present an interaction schematic display, which uses the same color code as the molecular graphics thumbnail. Interaction schematics are computed using the Graphviz library (see http://www.graphviz.org). The interaction schematic displays polypeptide molecules as circles, nucleic acids as squares and small molecules/chemicals as diamonds. Circles and squares may vary in size, as the schematics reflect differences in sequence length/molecule size. A line is drawn between two symbols if the two corresponding molecules were found to interact. Resting the mouse-pointer over a symbol will generate a pop-up showing the molecule name, and double-clicking on a symbol will scroll the page down and show the corresponding row in the table below the graphic images, which lists individual molecules and their interactions in detail. Table rows give counts of identical molecules in the biological unit, may summarize sequence annotation, and list the names of molecules found to interact. Sequence annotation summaries can be expanded to reveal interactive graphics that provides links to structure neighbors and annotations with conserved domains (7). Extensive and detailed help documentation is available by clicking on the question mark icons ‘?’ on the summary pages. Each icon ‘?’ is linked to the appropriate section in the help document.
As an example of the power of the integrated structure databases, assume we are interested in chemicals that may bind to the DNA-directed RNA polymerase II subunit Rpb1 from yeast, as shown in Figure 1. The first thing to try is the IBIS link for this protein, but in this instance there is only one protein-chemical binding site displayed. Feeling more adventurous, if we follow the VAST link for protein (chain) A, the first structure on the list is a RNA polymerase from Thermus thermophilus (PDB accession 3DXJ). If we follow the IBIS link for the 3DXJ structure we find there are seven antibiotics that bind to the protein chain D, which is structurally similar to the first yeast RNA polymerase. We note that the low level of sequence identity (24%) between the yeast RNA polymerase and the bacterial RNA polymerase is the reason the antibiotic binding sites did not show up in the first IBIS query, since IBIS uses a conservative threshold of ≥30% sequence identity for inferences.
RELATED STRUCTURES: CBLAST
CBLAST is a web service that visualizes similarities between proteins in NCBI's Entrez database [or arbitrary proteins submitted to NCBI's BLAST service (8)] and those with known 3D structures tracked in MMDB. It provides lists of homologous 3D structures for protein records in Entrez, as well as alignment details and visualization of sequence-structure alignments via the Cn3D viewer. Alignments between a query protein of interest and sequence-similar proteins with known 3D structure may serve as the starting point for the investigation of sequence–structure–function relationships or molecular modeling. CBLAST displays also provide information about conserved protein domains identified in the query proteins, which may provide a protein family context for the similar structures identified by the database search. A queuing schema has recently been developed for CBLAST, addressing database growth and an increase in user requests. In addition, the user interface and data presentation for CBLAST have also been updated and enhanced. The CBLAST home page provides a query box for users to directly enter GI numbers (‘GenBank Ids’, i.e. numerical sequence identifiers tracked by NCBI's Entrez system). If no GI number is available, the sequence may be submitted for a protein BLAST search, and links to CBLAST results are available on the formatted protein BLAST results. For all sequence records in the Entrez protein database, CBLAST results are pre-calculated and updated on a daily basis as new protein records or 3D structure data are made available, stored in a relational database, and can be instantly retrieved by following the ‘Related Information: Related Structure (Summary)’ link in the right margin of a protein sequence record display. CBLAST's pre-calculated sequence–structure alignments also support the IBIS service for inferring interaction sites and partners for proteins in Entrez.
SIMILAR STRUCTURES: VAST AND VAST SEARCH
NCBI continues to pre-compute results for all-against-all structure neighboring tethered to MMDB, using the VAST (9) algorithm. For analysis of structural similarities, protein 3D structures are parsed geometrically to reveal structurally compact subdomains. The search for structure neighbors is performed for such subdomains as well as for the whole molecule. Structurally compact subdomains, when present, are indicated in sequence annotation views as provided by the structure summary pages (Figure 1) and labeled ‘3d Domains’. On those sequence annotation views, the user can click on 3D domain segments to see a list of structure neighbors that contain a similar 3D domain, or click on the gray bar representing the full-length protein to see a list of structure neighbors that are similar in shape to all or a part of the whole molecule. Links to VAST 3D structure neighbors are also available via the ‘Similar Structures’ link provided at the top of the structure summary pages. Maintained together with the VAST database of pre-computed structure alignments and superpositions is VAST-Search, an interactive search service that lets users upload 3D coordinate sets in PDB file format, for structures not yet found in MMDB.
FUNDING
Funding to pay the open access charge: Intramural Research Program of the National Library of Medicine at National Institutes of Health/DHHS. Comments, suggestions and questions are welcome and should be directed to: info@ncbi.nlm.nih.gov.
Conflict of interest statement. None declared.
REFERENCES
- 1.Rose PW, Beran B, Bi C, Bluhm WF, Dimitropoulos D, Goodsell DS, Prlic A, Quesada M, Quinn GB, Westbrook JD, et al. The RCSB Protein Data Bank: redesigned web site and web services. Nucleic Acids Res. 2011;39:D392–D401. doi: 10.1093/nar/gkq1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 3.Wang Y, Addess KJ, Chen J, Geer LY, He J, He S, Lu S, Madej T, Marchler-Bauer A, et al. MMDB: annotating protein sequences with Entrez's 3D-structure database. Nucleic Acids Res. 2007;35:D298–D300. doi: 10.1093/nar/gkl952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shoemaker BA, Zhang D, Thangudu RR, Tyagi M, Fong JH, Marchler-Bauer A, Bryant SH, Madej T, Panchenko AR. Inferred Biomolecular Interaction Server—a web server to analyze and predict protein interacting partners and binding sites. Nucleic Acids Res. 2010;38:D518–D524. doi: 10.1093/nar/gkp842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang Y, Geer LY, Chappey C, Kans JA, Bryant SH. Cn3D: sequence and structure views for Entrez. Trends Biochem. Sci. 2000;25:300–302. doi: 10.1016/s0968-0004(00)01561-9. [DOI] [PubMed] [Google Scholar]
- 6.Sayle RA, Milner-White EJ. RASMOL: biomolecular graphics for all. Trends Biochem. Sci. 1995;20:374. doi: 10.1016/s0968-0004(00)89080-5. [DOI] [PubMed] [Google Scholar]
- 7.Marchler-Bauer A, Lu S, Anderson JB, Chitsaz F, Derbyshire MK, DeWeese-Scott C, Fong JH, Geer LY, Geer RC, Gonzales NR, et al. CDD: a Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res. 2011;39:D225–D229. doi: 10.1093/nar/gkq1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gibrat JF, Madej T, Bryant SH. Surprising similarities in structure comparison. Curr. Opin. Struct. Biol. 1996;6:377–385. doi: 10.1016/s0959-440x(96)80058-3. [DOI] [PubMed] [Google Scholar]