Abstract
The Mouse Genome Database (MGD, http://www.informatics.jax.org) is the international community resource for integrated genetic, genomic and biological data about the laboratory mouse. Data in MGD are obtained through loads from major data providers and experimental consortia, electronic submissions from laboratories and from the biomedical literature. MGD maintains a comprehensive, unified, non-redundant catalog of mouse genome features generated by distilling gene predictions from NCBI, Ensembl and VEGA. MGD serves as the authoritative source for the nomenclature of mouse genes, mutations, alleles and strains. MGD is the primary source for evidence-supported functional annotations for mouse genes and gene products using the Gene Ontology (GO). MGD provides full annotation of phenotypes and human disease associations for mouse models (genotypes) using terms from the Mammalian Phenotype Ontology and disease names from the Online Mendelian Inheritance in Man (OMIM) resource. MGD is freely accessible online through our website, where users can browse and search interactively, access data in bulk using Batch Query or BioMart, download data files or use our web services Application Programming Interface (API). Improvements to MGD include expanded genome feature classifications, inclusion of new mutant allele sets and phenotype associations and extensions of GO to include new relationships and a new stream of annotations via phylogenetic-based approaches.
INTRODUCTION
The Mouse Genome Database (MGD) (1–3) serves as a primary resource for mammalian biologists, delivering a spectrum of genetic, genomic and biological data supporting the use of mouse as a model for understanding human biology and disease. Central to its data offerings are the canonical mouse gene catalog, nucleotide and protein sequence associations, gene-to-function assignments based on the Gene Ontology (GO) (4), a comprehensive catalog of mutant alleles, associations of mutant genotypes to their phenotype through the Mammalian Phenotype (MP) Ontology (5) and to the human diseases for which they are a model through curated associations to human diseases in Online Mendelian Inheritance in Man database (OMIM) (6). In addition, MGD provides a comprehensive genetic map, a genome browser (Mouse GBrowse) for genome viewing, Single Nucleotide Polymorphisms (SNPs) and other polymorphisms and mammalian orthology data. A summary of the current contents of MGD is given in Table 1.
Table 1.
Summary of MGD data content (14 September 2011)
| Genes with nucleotide sequence data | 28 803 |
| Genes with protein sequence data | 25 070 |
| Genes with mutant alleles in mice | 15 145 |
| Genes with one of more mutant allelesa | 20 397 |
| Total mutant allelesa | 738 414 |
| Number of cre-containing transgenes and knock-ins | 1511 |
| Genes with mouse experiment-based functional (GO) annotations | 13 524 |
| Mouse/human orthologs | 17 847 |
| Mouse/rat orthologs | 16 686 |
| Human diseases with one or more mouse models | 1121 |
| QTLs | 4670 |
| Number of references | 169 700 |
| Number of reference SNPs | 10 089 892 |
aMutant alleles include those occurring in mice and those existing only in mouse ES cell lines. Of the 738 414 total mutant alleles, 682 745 are gene traps in ES cell lines.
Integrated with MGD are other components of the Mouse Genome Informatics (MGI) database resource (http://www.informatics.jax.org). These include the Gene Expression Database (7), the Mouse Tumor Biology Database (8) and the MouseCyc database of metabolic pathways (9). Two additional resources tied to the main MGI resource are the International Mouse Strain Resource (IMSR) (10) and the Recombinase (cre) Portal (1).
Data in MGD are obtained through data loads from major resource providers [e.g. sequence data from GenBank, gene models from NCBI, Ensembl, VEGA, mutant alleles from N-ethyl-N-nitrosourea (ENU)-mutagenesis groups and International Knockout Mouse Consortium (IKMC)], from electronic submissions from investigator laboratories, and from the biomedical literature. All data are attributed to the original source with access to references provided via PubMed where available. For data loads, quality control reports are generated that enumerate format and/or content anomalies and prioritize errors that need attention by curators. Standards for gene, allele and strain nomenclature, and for functional, phenotypic and human disease annotations using vocabularies and ontologies enable consistent annotations and robust data retrieval.
MGD data can be accessed in many ways. A Quick Search box appears on all web pages and provides a ubiquitous, fast and simple entry for broad keyword or ID searches. More specialized query forms, accessible via the Search pull down on the navigation bar, allow multiparameter advanced searches, and the data content area icons on the homepage lead users to specific accesses to that data area. A vocabulary browser supports access to MGD content through ontology terms. A variety of regularly updated database reports can be accessed on the File Transfer Protocol (FTP) site. Programmatic access is provided through web services and through direct SQL access.
KEY UPDATES AND CHANGES IN 2011
Expanded classification terms for genome features
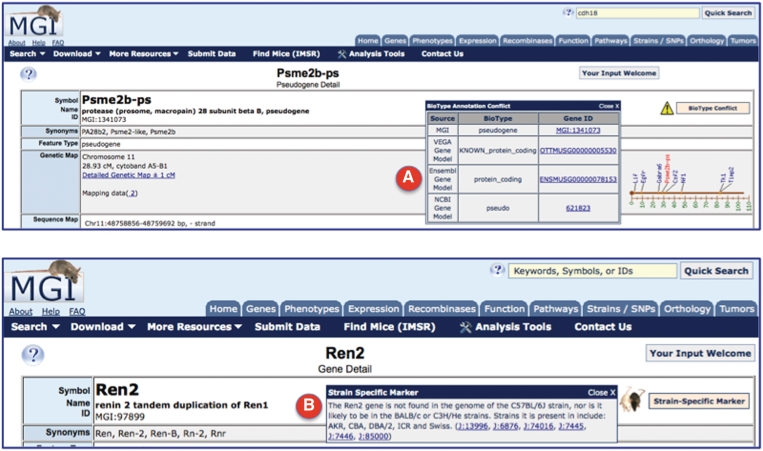
New to MGD are feature type classifications as attributes of genome features. The feature types allow users to refine searches to include only specific classes of genome features (protein-coding genes, mircoRNAs, lincRNAs, Quantitative Trait Loci (QTL), transgenes, pseudogenes, etc.). Most of the classification terms and definitions are derived from the Sequence Ontology (SO) (11). We have also added new subclassification terms for genome features formerly grouped as pseudogenes. The overarching term for these genome features is now pseudogenic region (SO: 0000462), defined as a non-functional feature descended from a gene or other functional feature. In MGD, three subcomponents: pseudogene (a sequence that closely resembles a known functional gene, at another locus within a genome, which is non-functional as a consequence of mutations that prevent its transcription or translation); pseudogenic gene segment (a recombinational unit of a gene which when incorporated by somatic recombination in the final gene transcript result in a non-functional product); and polymorphic pseudogene (a pseudogene lacking function owing to a SNP or deletion/insertion, but in other individuals/haplotypes/strains the gene is translated) are currently in use. Where MGD, VEGA, Ensembl and National Center for Biotechnology Information (NCBI) disagree on the pseudogene subclassification type, a biotype conflict note is presented to the user on the MGD locus detail page. Where a genome feature is a non-functional pseudogene in some mouse strains, but functional in other mouse strains, a strain-specific note is presented on the detail page (Figure 1).
Figure 1.
Screenshots of the upper portion of two locus detail pages. (A) The BioType Conflict indicator (upper right), when opened, displays the different biotype annotations for Psme2b-ps. In this case, MGI and NCBI assign this marker as a pseudogene, where VEGA and Ensembl have assigned the status as protein coding gene. Links are provided to the underlying evidence that support the biotype assignments by different annotation groups. (B) The strain-specific marker indicator (upper right), when opened, displays information about strains in which the gene (in this case Ren2) is found or, not found, in the genome, with supporting reference links.
Nomenclature harmonization: T-cell receptor and immunoglobulin gene segments
Working with the Immunogenetics Information System, IMGT/Gene-DB (12), MGD has expanded the number of defined T-cell receptor and immunoglobulin gene segments (a gene component region which acts as a recombinational unit of a gene whose functional form is generated through somatic recombination) to over 670 and harmonized nomenclature for these important immunological gene segments.
Mutant allele sets added
The number of mutant alleles in MGD has increased by over 23 640 this year. This largely reflects ongoing development of genetically engineered and ENU-induced mutations by major mutagenesis programs, with significant contributions by individual investigators. Among the major additions of new mutant alleles to MGD were: 8364 new targeted mutations added from the IKMC (13), 870 new transgenes added from the Gene Expression Nervous System Atlas project (14), 492 new targeted and gene trap mutations from a Genentech/Lexicon collaboration (15) and over 200 new ENU mutations from Dr Bruce Beutler's Mutagenetix program (16). Over 3000 new mutant alleles were developed from investigator-initiated experiments and added to MGD from biomedical literature curation or via investigator data submissions to MGD. The remaining approximately 10 000 new alleles are gene traps added via a data load from NCBI's Genome Survey Sequences Database (GSS) (17), most of which were generated by the IKMC. Of the current more than 596 000 mutant alleles for mice, most were generated and only exist in Embryonic Stem (ES) cell lines, with approximately 30 400 of these being either created or developed into living mice.
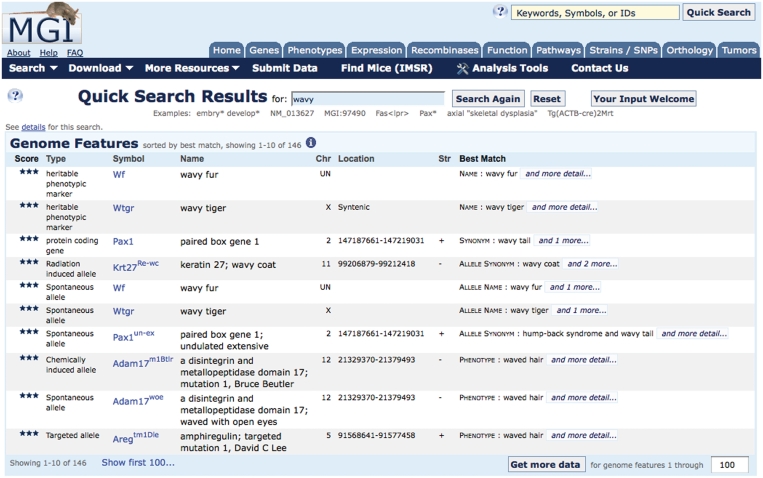
The Quick Search tool now includes mutant alleles
To take advantage of the large number of new mutant allele resources, MGD has improved the characteristics its Quick Search tool, so it now returns the alleles, as well as other genome features, most closely associated with a query. (The previous implementation of the Quick Search returned genome features at the level of the gene.) This change helps users more easily locate relevant mouse model data from queries for phenotypes or disease. Given that there are Quick Search accounts for >90% of the interactive MGD searches, we expect this change to have significant beneficial impact (Figure 2).
Figure 2.
Screenshot of the results of querying for ‘wavy’ using the MGI Quick Search box. Note that heritable phenotypic markers that identify mutants whose underlying gene is not yet identified, such as Wf and Wtgr are retrieved, as well as genes (e.g. Pax1, with synonym of wavy tail), and other types of mutant alleles in defined genes (e.g. Pax1un-ex, undulated extensive mutation of the Pax1 gene).
Extensions to GO annotations
GO annotations are being extended via phylogenetic-based approaches. Through identification of phylogenetically related orthologous, homologous and paralogous genes across species, the GO consortium is promoting coordinate annotations of these genes across organisms. MGD is actively participating in these gene annotations to enrich functional information about a highly curated set of phylogenetically related genes among species and to enable propagation of functional annotations between organisms (18,19).
Retooling MGD infrastructure: a plan for the future
MGD is in the process of a significant infrastructure migration project to move from the Sybase relational database management system to a more technically attractive open source database technology (PostgreSQL). Phase I of this project is to move and rewrite software on our public servers, specifically those components supporting the web interface and direct SQL accounts. As well, we are retooling the web interface software to use Solr and Lucene to handle most querying, Java Spring Model-View-Controller (MVC) for web page generation and YAHOO User Interface (YUI) for on-page interactivity. Beyond the user benefits visible in the initial release, this technology migration will position us well for future developments. Phase II, to migrate and retool the software residing on our back end servers (where the data loading and curation occur) is also underway.
New direct access methods for MGD
MGD has always provided direct SQL access to a public Sybase server. As part of the migration described in the previous paragraph, the Sybase server has been retired, and a public PostgreSQL server is now available. In addition, for users who want MGD at their local sites, we now provide complete database dumps for both PostgreSQL and MySQL. The public SQL server and the database dumps are updated on a weekly basis. Dump files are available from our FTP site at ftp://ftp.informatics.jax.org/pub/database_backups/. Instructions can be found at http://www.informatics.jax.org/software.shtml. Contact MGI User Support (mgi-help@informatics.jax.org) to request a PostgreSQL account or for assistance in using the database dumps. Individuals interested in programmatic and bulk access may also want to join the MGI-Technical listserve (http://www.informatics.jax.org/mgihome/lists/lists.shtml) to receive technical updates about the database.
OTHER INFORMATION
Mouse gene, allele and strain nomenclature
MGD is the international authoritative source of symbols and names for mouse genes, alleles and strains. MGD follows and implements the guidelines set by the International Committee on Standardized Genetic Nomenclature for Mice (http://www.informatics.jax.org/nomen). This official nomenclature is widely disseminated through regular data exchange and curation of shared links between MGI and other bioinformatics resources. MGD staff members work with editors of journal publications and consortium projects to promote adherence to mouse nomenclature standards in publications and online data resources.
To support consistency of nomenclature across species, MGD coordinates names and symbols for genes and genome features with nomenclature experts from the Human Gene Nomenclature Committee (HGNC) (20) (http://www.genenames.org/) and the Rat Genome Database (RGD) (21) http://rgd.mcw.edu). The MGD nomenclature coordinator can be contacted by email (nomen@informatics.jax.org).
Programmatic and bulk data access
Portions of the database are accessible programmatically using web services and BioMart. The MGI web service accepts SOAP 1.1 and 1.2 requests. For details, see http://www.informatics.jax.org/mgihome/other/web_service.shtml. The MGD BioMart is accessible at http://biomart.informatics.jax.org. Additional information about MartServices can be found at http://www.biomart.org/martservice.html.
MGI also provides bulk data sets through regularly updated FTP reports (ftp://ftp.informatics.jax.org/pub/reports/index.html) and via the MGI Batch Query tool (http://www.informatics.jax.org/javawi2/servlet/WIFetch?page=batchQF) where users can develop a customized bulk data set.
Electronic data submission
MGD accepts contributed data sets from individuals and organizations for any type of data maintained by the database. The most frequent types of contributed data are mutant and phenotypic allele information originating with the large mouse mutagenesis centers and strain data from repositories that contribute to the IMSR (http://www.findmice.org) (10). Each electronic submission receives a permanent database accession ID. All data sets are associated with their source, either a publication or an electronic submission reference. Details about data submission procedures can be found at http://www.informatics.jax.org/submit.shtml.
Additions and corrections to the representation of data and information in MGD can be submitted using the ‘Your Input Welcome’ link that appears in the upper right hand corner of gene and allele detail pages.
Community outreach and User Support
The MGD resource has full time staff members who are dedicated to user support and training. Members of the User Support team can be contacted via email, web requests, phone or Fax.
World wide web: http://www.informatics.jax.org/mgihome/support/support.shtml
Email access: mgi-help@informatics.jax.org
Telephone access: 1 207 288 6445
Fax access: 1 207 288 6132
MGD User Support staff are available for on-site training on the use of MGD and other MGI data resources. MGD's traveling tutorial program (roadshow) includes lectures, demos and hands-on tutorials, which can be customized according to the research interests of the audience. To inquire about sponsoring a MGD roadshow, send email to mgi-help@informatics.jax.org.
On-line training materials for MGD and other MGI data resources are available as FAQs and on-demand help documents.
Other outreach
MGI-LIST (http://www.informatics.jax.org/mgihome/lists/lists.shtml) is a moderated and active email bulletin board supported by the MGD User Support group. The MGI listserve has over 2100 subscribers. On average, there are three posts per day, every day. The MGI-Technical listserve also has been instituted for technical information for software developers and bioinformaticians accessing MGI data, using APIs, and making links to MGI.
HIGH LEVEL OVERVIEW OF THE MAIN COMPONENTS AND IMPLEMENTATION
The MGD production database comprises approximately 180 tables within which biological information is encoded. As we are transitioning between database engines, we currently have instances in both Sybase and PostgreSQL. BLAST-able databases, genome assembly files for sequence data and images are stored outside the relational database. An editing interface and automated load programs are used to input data into the MGD system. Automated loads enter/update the bulk of data and associations in MGD. A typical load will load ‘as much as it can’(typically, the large majority) and report the rest in various quality control reports. These are reviewed by curators, who may resolve problem cases by editing MGD and/or by communicating with data providers. The interactive graphical editing interface provides curators with the ability to update the database, enter new data from the literature, track curation status, etc.
Public data access to MGD is provided primarily through the web interface where users can interactively query and download our data through a web browser. MouseBLAST allows users to do sequence similarity searches against a variety of rodent sequence databases that are updated weekly from selected sequence databases from NCBI, UniProt and other providers. Mouse GBrowse allows users to visualize mouse data sets against the genome as a series of linear tracks. All MGD files and programs are openly and freely available.
We continue to provide MGD BioMart with the addition of new classification terms for genome features. MGD BioMart supports chaining to several other BioMarts including Ensembl, VEGA and RGD. Additional functionalities such as the ability to filter by GO, MP Ontology and OMIM terms, and including additional information about alleles, are planned for future extensions. MGD BioMart is updated on a weekly basis.
CITING MGD
For a general citation of the MGI resource please cite this article. In addition, the following citation format is suggested when referring to data sets specific to the MGD component of MGI: MGD, MGI, The Jackson Laboratory, Bar Harbor, Maine (URL: http://www.informatics.jax.org). [Type in date (month, year) when you retrieved the data cited.].
FUNDING
National Institutes of Health/National Human Genome Research Institute, The Mouse Genome Database (grant HG000330). Funding for open access charge: National Institutes of Health/ NHGRI (grant HG000330).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
M. T. Airey, A. Anagnostopoulos, R. Babiuk, R. M. Baldarelli, J. S. Beal, S. M. Bello, N. E. Butler, J. Campbell, L. E. Corbani, S. L. Giannatto, H. Dene, M. E. Dolan, H. R. Drabkin, K. L. Forthofer, M. Knowlton, J. R. Lewis, M. McAndrews-Hill, S. McClatchy, D. S. Miers, L. Ni, H. Onda, J. E. Ormsby, J. M. Recla, D. J. Reed, B. Richards-Smith, D. R. Shaw, R. Sinclair, D. Sitnikov, C. L. Smith, M. Tomczuk, L. L. Washburn, Y. Zhu.
REFERENCES
- 1.Blake JA, Bult CJ, Kadin JA, Richardson JE, Eppig JT, the Mouse Genome Database Group The Mouse Genome Database (MGD): premier model organism resource for mammalian genomics and genetics. Nucleic Acids Res. 2011;39:D842–D848. doi: 10.1093/nar/gkq1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bult CJ, Kadin JA, Richardson JE, Blake JA, Eppig JT, the Mouse Genome Database Group The Mouse Genome Database: enhancements and updates. Nucleic Acids Res. 2010;38:D536–D592. doi: 10.1093/nar/gkp880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blake JA, Bult CJ, Eppig JT, Kadin JA, Richardson JE, the Mouse Genome Database Group The Mouse Genome Database genotypes: phenotypes. Nucleic Acids Res. 2009;37:D712–D719. doi: 10.1093/nar/gkn886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.The Gene Ontology Consortium. The Gene Ontology in 2010: extensions and refinements. Nucleic Acids Res. 2010;38:D331–D335. doi: 10.1093/nar/gkp1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Smith CL, Eppig J. The mammalian phenotype Ontology: enabling robust annotation and comparative analysis. Wiley Interdiscip. Rev. Syst. Biol. Med. 2009;1:390–399. doi: 10.1002/wsbm.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Amberger J, Bocchini C, Hamosh A. A new face and new challenges for Online Mendelian Inheritance in Man (OMIM®) Hum. Mutat. 2011;32:564–567. doi: 10.1002/humu.21466. [DOI] [PubMed] [Google Scholar]
- 7.Finger JH, Smith CM, Hayamizu TF, McCright IJ, Eppig JT, Kadin JA, Richardson JE, Ringwald M. The mouse Gene Expression Database (GXD): 2011 update. Nucleic Acids Res. 2011;39:D835–D841. doi: 10.1093/nar/gkq1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Begley DA, Krupke DM, Neuhauser SB, Richardson JE, Bult CJ, Eppig JT, Sundberg JP. The Mouse Tumor Biology Database (MTB): a central electronic resource for locating and integrating mouse tumor pathology data. Vet. Pathol. 2011 doi: 10.1177/0300985810395726. January 31 (doi: 10.1177/0300985810395726; epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Evsikov AV, Dolan ME, Genrich MP, Pated E, Bult CJ. MouseCyc: a curated biochemical pathways database for the laboratory mouse. Genome Biol. 2009;10:R84. doi: 10.1186/gb-2009-10-8-r84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Strivens M, Eppig JT. Visualizing the laboratory mouse: capturing phenotypic information. Genetica. 2004;122:89–97. doi: 10.1007/s10709-004-1435-7. [DOI] [PubMed] [Google Scholar]
- 11.Eilbeck K, Lewis SE, Mungall CJ, Yandell M, Stein L, Durbin R, Ashburner M. The Sequence Ontology: a tool for the unification of genome annotations. Genome Biol. 2005;6:R44. doi: 10.1186/gb-2005-6-5-r44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lefranc MP, Giudicelli V, Ginestoux C, Jabado-Michaloud J, Folch G, Bellahcene F, Wu Y, Gemrot E, Brochet X, Lane J, et al. IMGT, the international ImMunoGeneTics information system. Nucleic Acids Res. 2009;37:D1006–D1012. doi: 10.1093/nar/gkn838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ringwald M, Iyer V, Mason J, Stone K, Tadepally H, Kadin JA, Bult CJ, Eppig JT, Oakley D, Briois S, et al. The IKMC Web Portal: a central point of entry to data and resources from the International Knockout Mouse Consortium. Nucleic Acids Res. 2011;39:D849–D855. doi: 10.1093/nar/gkq879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gong S, Kus L, Heintz N. Rapid bacterial artificial chromosome modification for large-scale mouse transgenesis. Nat. Protoc. 2010;5:1678–1696. doi: 10.1038/nprot.2010.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tang T, Li L, Tang J, Li Y, Lin WY, Martin F, Grant D, Solloway M, Parker L, Ye W, et al. A mouse knockout library for secreted and transmembrane proteins. Nat. Biotechnol. 2010;28:749–755. doi: 10.1038/nbt.1644. [DOI] [PubMed] [Google Scholar]
- 16.Hoebe K, Beutler B. Unraveling innate immunity using large scale N-ethyl-N-nitrosourea mutagenesis. Tissue Antigens. 2005;65:395–401. doi: 10.1111/j.1399-0039.2005.00369.x. [DOI] [PubMed] [Google Scholar]
- 17.Sayers EW, Barrett T, Benson DA, Bolton E, Bryant SH, Canese K, Chetvernin V, Church DM, DiCuccio M, Federhen S, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2011;39:D38–D51. doi: 10.1093/nar/gkq1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gaudet P, Livstone MS, Lewis SE, Thomas PD. Phylogenetic-based propagation of functional annotations within the Gene Ontology consortium. Brief. Bioinform. 2011;12:449–462. doi: 10.1093/bib/bbr042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.The Reference Genome Group of the Gene Ontology Consortium. The Gene Ontology's Reference Genome Project: a unified framework for functional annotation across species. PLoS Comp. Biol. 2009;5:e1000431. doi: 10.1371/journal.pcbi.1000431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Seal R, Gordon S, Lush M, Bruford E, Wright M. genenames.org: the HGNC resources in 2011. Nucleic Acids Res. 2011;39:D514–D519. doi: 10.1093/nar/gkq892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dwinell M, Worthey EA, Shimoyama M, Bakir-Gungor B, DePons J, Laulederkind S, Lowry T, Nigram R, Petri V, Smith J, et al. RGD Team The Rat Genome Database 2009: variation, ontologies and pathways. Nucleic Acids Res. 2009;37:D744–D749. doi: 10.1093/nar/gkn842. [DOI] [PMC free article] [PubMed] [Google Scholar]