Abstract
SNPedia (http://www.SNPedia.com) is a wiki resource of the functional consequences of human genetic variation as published in peer-reviewed studies. Online since 2006 and freely available for personal use, SNPedia has focused on the medical, phenotypic and genealogical associations of single nucleotide polymorphisms. Entries are formatted to allow associations to be assigned to single genotypes as well as sets of genotypes (genosets). In this article, we discuss the growth of this resource and its use by affiliated software to create personal genome reports.
INTRODUCTION
The promise of the human genome project includes the potential to know which variants in a person's genome are of medical significance. Information about variants are generally published in individual peer-reviewed publications in textual form, intended to be read by fellow researchers and the public, whereas a person's genomic information is stored in a large-scale data format that is far more computable than legible.
One way to bridge this gap is to convert the information presented in the literature into a machine-readable form. With the advent a decade ago of large-scale genomic microarray experiments, studies such as genome-wide association studies (GWAS) became possible.
SNPedia (pronounced i/snpi:di.ə/ SNIP-pee-dee-ə) is a wiki resource focusing on converting the significant information in large-scale genomic studies into a form that is legible to both researchers and programs. Online for 5 years, SNPedia summarizes the medical, phenotypic, forensic and genealogical associations of DNA variations. Entries are formatted in a systematic manner allowing such associations to be assigned to single genotypes as well as sets of genotypes (genosets). In this article, we discuss the growth of this resource and its use by affiliated software to create personal genome reports. SNPedia is freely available for personal use at http://www.SNPedia.com/.
INFORMATION COLLECTION AND CONTENT
Wiki format
SNPedia is a Semantic MediaWiki site (1) that is edited and updated by both automatic and manual means. It is intended to be formatted in a manner supporting automated report analysis and generation by associated software (such as Promethease, discussed below), while still retaining a level of legibility for both casual and frequent users.
Rs numbers
The NCBI dbSNP database (2) catalogs over 10 million non-redundant Reference SNPs. Each is assigned an identifier which begins with the two-letter code rs and then a unique number. This identifier is in widespread use, such as on large-scale microarrays, and is now common in scientific literature. It also became accessible to non-scientists via Direct To Consumer (DTC) genotyping services. The rs# identifier provides a precisely defined location in the genome and is easily parsed from the scientific literature. It is stable across genome builds, so it does not require researchers to periodically remap old coordinates onto newer builds. While most commonly representing a single nucleotide polymorphism (SNP), dbSNP is not limited to single nucleotide variants and thus an rs# may also represent indels of varying size. For example, rs333 is an indel of 32 nt. In contrast, dbVAR (2) covers larger variants. In terms of species coverage, while dbSNP is species agnostic, SNPedia focuses on human data at this time but does contain some non-human rs#s to ensure future support.
Types of content
The basic units of information in SNPedia are single or multiple nucleotide positions in the human genome known to vary in either germline or somatic contexts. SNPedia currently collects information on single nucleotide variants such as SNPs and mutations, or more specifically, on genotypes composed of one or more variant loci. A page of content is created based on a single rs-identifier (as maintained in dbSNP), and then typically three associated pages are created, reflecting the three possible genotypes for that SNP (homozygous for the major allele, heterozygous and homozygous for the minor allele at that locus). To the extent possible, a summary of the odds ratios for one or more associated medical conditions is reported for each genotype. Sets of genotypes from unlinked loci, known as genosets, are also defined in SNPedia. Additional content types include the genes, phenotypes, medical conditions, and drug interactions reported to be associated with these variants.
Users contribute both structured data and free text comments using a combination of standard Mediawiki syntax and Semantic Forms. Example properties include ‘Magnitude’, which gives a qualitative summary of significance on a 0.0–10 scale, and ‘Repute’ for consequences that can be classified as clearly ‘Good’ or ‘Bad’. Unlimited wiki text with hyperlinks, images and formatting provides the ability to communicate subtlety lacking from more structured data formats.
Sources of content
Data is collected from both bulk and individual sources. As a wiki, users add data on a continuous basis, and these additions are augmented by periodic updates text mined from public data sources. Sources of content cite publications, in particular, Pubmed PMID or DOI identifiers. SNPedia is committed to maintaining free access for personal users to all contributed information.
Criteria for inclusion
While SNPedia casts a broad net with regard to the creation of ‘rs-pages’ defining individual SNPs, genotype-specific pages are primarily only created for variants that have significant medical or genealogical consequences based on published meta-analyses, studies of at least 500 patients or two or more independent studies (i.e. replicated findings), or other historic, statistical or medical significance. This allows software creating personal genome reports, which is based on the genotypes carried with an individual, to create more robust reports. With some exceptions, SNPedia's genotype-specific pages generally do not include variants that are unreplicated or from studies with less statistical power. However, variants with high penetrance, such as ones that might also be reported in OMIM (3) or in LSDBs, are increasingly being added to SNPedia. Often these variants are so infrequent that they have not been observed in any populations sampled for variation, and therefore they are not present in dbSNP. In such cases, SNPedia submits their genomic data directly to dbSNP in order to have rs numbers assigned. After release in dbSNP, these variants are then added to SNPedia. An example of such a variant is the del-F508 mutation representing the most common cystic fibrosis-causing variant (4), now represented in dbSNP (and therefore SNPedia) as rs113993960. Anyone interested in having specific variants added to dbSNP in this fashion is encouraged to contact SNPedia.
Data curation
There are at least four levels of data curation. First, all additions and changes to the SNPedia are reviewed by wiki users (including the editors). Second, Semantic Mediawiki templates flag certain warnings (such as SNPs with no assigned chromosome, or known by older/expired names) to bring them prominently to the readers and editors attention. Third, at least two independently developed software bots frequently crawl the entire site from the outside. They add supplementary information such as chromosome, position, gene and allelic data. While designed to be cautious and not replace any information entered by a human, these bots often are the first to detect irregularities. Fourth, SNPedia content is used in Promethease personal genome reports read by a diverse audience able to recognize problematic assertions. They are uniquely able and motivated to detect and report errors or nuances missed in the original research.
Content growth
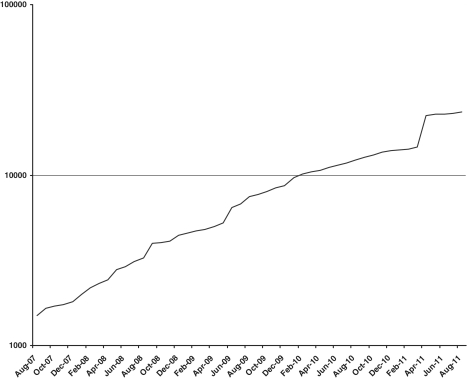
The SNPedia wiki has been accessible online since 2006, pre-dating the advent of the DTC genomic testing companies. Starting from under 1000 SNPs, it now has approximately 25 000 SNPs (Figure 1), 10 000 genotypes and 200 genosets. SNPs are associated with 200 or more medical conditions and 150 or more drugs. More importantly, the growth in both new entries per week as well as in edits per page over the last 5 years has been steady and consistent. Additional statistics that are tracked include the number of SNPs associated with each disease category (for the 10 with the highest number of associated SNPs, the range is from 114 to 229; for all diseases, the average is 27 SNPs/condition), and the number of Pubmed PMIDs per SNP (currently ranging from 29 to 60 for the 10 SNPs with the highest number of associated references).
Figure 1.
SNPedia Growth. The total number of individual SNPs described in SNPedia for the last 4 years. Note that the y-axis is logarithmic.
Genosets
Many interesting phenotypes are dependent on more than a single variant. As mentioned above, in order to accurately model this, SNPedia has introduced a notation for such sets of genomic variants, known as ‘genosets’, and implemented a parser in Promethease. For example, to recognize the two SNPs which define homozygosity for APOE4 the genoset criteria are noted exactly as follows:
and(rs429358(C;C),rs7412(C;C))
and the possibility of having at least one APOE4 allele is represented as:
and(rs429358(C),rs7412(C))
Genosets can refer to other genosets, and cyclical references are resolved. The boolean operators (‘and’, ‘or’ ‘not’) as well as ‘atleast(N, list)’ are also supported by Promethease. The present nomenclature makes it difficult to distinguish linked SNPs from the same strand (i.e. in cis), which while appropriate for data from microarrays and from unlinked loci, will require enhancing given the advent of full genome sequencing and phased data.
FUNCTIONALITY
Automated data gathering
Automated software gathers much of the base rs# page information in SNPedia. The bulk of information in SNPedia is gathered by software which checks Pubmed daily. It identifies papers which mention an rs# in their abstract, and it adds the title, PMID and a link to the paper to the appropriate rs# page in SNPedia. Other bots do weekly monitoring of Genome.gov, OMIM, PharmGKB and other websites of interest. Whether via addition by automatic or human editing, newly added rs#s get a second round of bot edits which update the chromosome, position, HUGO gene name, known alleles, HapMap population frequencies, microarray coverage and other relevant information.
Data availability
The information in SNPedia is available under a Creative Commons Attribution-Noncommercial-Share Alike 3.0 United States License and also by commercial license. In addition to its wiki web browser interface SNPedia is also available in Distributed Annotation System (DAS) (5) service form from The European Bioinformatics Institute http://www.ebi.ac.uk/das-srv/easydas/bernat/das/SNPedia or in GFF3 at http://www.snpedia.com/files/gbrowse/SNPedia.gff for use with GBrowse (6). The server also understands a Mediawiki API which answers requests in common formats (JSON, XML) and can be accessed with popular libraries in Perl, Python, Java and other languages.
External links
Each variant page includes a sidebar with a large number of external links. These links include links to the specific pages in both public genome databases (such as dbSNP, HapMap, Ensembl, PharmGKB) as well as selected private sources (such as NextBio and 23andMe). Typically, data from structured data sources such as the NIH/GWAS catalog and PharmGKB are presented in another sidebar also presented lower on the page.
Other features
What links here
Several wiki features can be particularly useful to a user of SNPedia. One example is the ‘What Links Here’ link found in the left navigation column; a click on this link from a drug, medical condition or gene page will provide a list of all content—and thus primarily SNP pages—that link to that page. While SNPs may already be mentioned in the page, this wiki feature provides a simple way to dynamically tabulate all associated SNPs at any given time.
Discussions
Another feature implemented in this wiki is the Discussion tab. Users may post a question or start a discussion on the Discussion page associated with any SNP page, often thereby aiding data curation efforts. An additional tab present on most pages is ‘Edit with Form’, which can be especially useful to users less familiar with the structured fields in use. Lastly, a search tool provides a simple means for quickly searching the entire wiki.
Promethease
A downloadable companion program to SNPedia, called Promethease, is available for both PC and Mac operating systems. Promethease is a free tool that builds a personal report based on a file of genotypes and the information in SNPedia at the time the program is run. For confidentiality reasons, Promethease runs on a user's PC and does not transfer a user's genotype to any central server. Customers of DTC or other genomic testing services that allow users access to their own bulk data (including 23andMe, FamilyTreeDNA, Complete Genomics and others) can routinely use Promethease to provide a report that is up to date and independent of the company that produced the genotype or sequence data. Promethease can also pool the data from multiple testing services, and has a variety of other features allowing comparisons between single or multiple individuals, as well as reporting carrier status and pharmacogenomic associations. The electronic report is completely hyperlinked to SNPedia, allowing users to directly evaluate the primary sources for all information.
Full genomes
SNPedia's information is effectively monitored and thus curated by individuals looking at their own DTC genomic data, and regardless of the data source (DNA chip or sequence) SNPedia users may publicly post their data online; full genomes are online at http://www.snpedia.com/index.php/Genomes. Not surprisingly, significantly more genome annotation and analysis is possible for full genomic sequence compared to DNA chip data.
For example, one participant of the Personal Genome Project (7) has received his full genome as sequenced by Complete Genomics, and additionally has been genotyped on multiple microarray platforms. As of 11 August 2011, SNPedia contained information on 23 987 SNPs, of which Promethease was able to annotate 19 972 genotypes from this participant's full genome data. Had he been genotyped on 23andMe's latest DTC platform with 100% efficiency, a maximum of 14 082 genotypes could have been analyzed, and his actual data from the older v2 platform provided only 12 123 genotypes.
This data set also presents another example of the utility of using full genome sequence in conjunction with SNPedia. While some individuals will be able to correctly detect carrier status (8), other variants tagged as lethal (or early-onset) and disease causing based on historic publications can readily be seen as ‘false positives’ when viewed in the context of a medical history free of the corresponding disease. In this case, the participant's genotype is one homozygous for a variant published as leading to Gaucher's disease. As more full genomes become available it is likely that this variant and many others (9) will be annotated as less pathogenic than indicated in the scientific literature. As this new information becomes available, updates are made to SNPedia and therefore also propagate to Promethease reports.
Self-reported phenotypes
SNPedia does not maintain Wikipedia's ‘No original research’ policy (http://en.wikipedia.org/wiki/Wikipedia:No_original_research), and instead actively invites well documented original research, as do certain DTC testing companies (10). One example with forensic application involves the genotypes influencing eye color. SNPedia's wiki format allows users to upload a picture of their eyes and record their genotypes and contact information into a shared online spreadsheet. This is useful for capturing complex phenotypes where an ontology has yet to be established.
FUTURE DIRECTIONS
The advent of large numbers of fully sequenced human genomes offers several changes that SNPedia will adapt to and incorporate. First, as sequence data for many more individuals becomes available, it will be possible to determine which of the variants reported in legacy variant databases are truly likely to be highly penetrant and consequential, and which are historic artifacts. SNPedia has added confirmed variants into dbSNP and will increase it's coverage of the highly penetrant (even if rare) variants as they are confirmed. Additionally, as sequence data supplants microarray data as a source of large-scale data for an individual, SNPedia will modify scripts and data structures, such as adding chromosomal strand-specific genosets. Second, as the nomenclature in publications becomes standardized for structural as well as methylated variants, there will be increased coverage of such variants (11). And lastly, it's likely that as connections between databases and wikis covering linked information become more robust, meta-efforts tying together the various—omics fields with phenotypic, pharmacogenomic and medical history information will become not only possible but actionable. Real-time information on overall wiki statistics for SNPedia are at URL http://www.snpedia.com/index.php/Special:Statistics, with the more salient (yet still real-time) statistics about the number of SNPs and related information summarized in the FAQ page at URL http://www.snpedia.com/index.php/SNPedia:FAQ/.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Materials 1 and 2.
FUNDING
Funding for open access charge: Waived by Oxford University Press.
Conflict of interest statement. The authors are the co-founders of River Road Bio, of which SNPedia and Promethease are wholly owned products.
ACKNOWLEDGEMENTS
The authors gratefully thank and acknowledge all our contributors, especially users James Lick, Kay and Meightysix.
REFERENCES
- 1.Krötzsch M, Vrandecic D, Völkel M, Haller H, Studer R. Semantic Wikipedia. J. Web Semant. 2007;5:251–261. [Google Scholar]
- 2.Sayers E, Barrett T, Benson D, Bolton E, Bryant S, Canese K, Chetvernin V, Church D, DiCuccio M, Federhen S, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2011;39:D38–D51. doi: 10.1093/nar/gkq1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hamosh A, Scott A, Amberger J, Bocchini C, McKusick V. Online Mendelian Inheritance in Man (OMIM), a knowledgebase of human genes and genetic disorders. Nucleic Acids Res. 2005;33:D514–D517. doi: 10.1093/nar/gki033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Riordan J, Rommens J, Kerem B, Alon N, Rozmahel R, Grzelczak Z, Zielenski J, Lok S, Plavsic N, Chou J, et al. Identification of the cystic fibrosis gene: cloning and characterization of complementary DNA. Science. 1989;245:1066–1073. doi: 10.1126/science.2475911. [DOI] [PubMed] [Google Scholar]
- 5.Prlic A, Down T, Kulesha E, Finn R, Kahari A, Hubbard T. Integrating sequence and structural biology with DAS. BMC Bioinformatics. 2007;8:333. doi: 10.1186/1471-2105-8-333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stein L, Mungall C, Shu S, Caudy M, Mangone M, Day A, Nickerson E, Stajich E, Harris TW, Arva A, et al. The generic genome browser: a building block for a model organism system database. Genome Res. 2002;12:1599–1610. doi: 10.1101/gr.403602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Church G. The personal genome project. Mol. Sys. Biol. 2005;1:30. doi: 10.1038/msb4100040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Roberts M, Riegert-Johnson D, Thomas B. Self diagnosis of lynch syndrome using direct to consumer genetic testing: a case study. J. Genet. Counsel. 2011;20:327–329. doi: 10.1007/s10897-011-9356-y. [DOI] [PubMed] [Google Scholar]
- 9.Davies K. Jay talking personal genomes. Bio-IT World. 2010 http://www.bio-itworld.com/BioIT_Article.aspx?id=101667 (6 September, date last accessed) [Google Scholar]
- 10.Do C, Tung J, Dorfman E, Kiefer A, Drabant E, Francke U, Mountain J, Goldman S, Tanner C, Langston J, et al. Web-based genome-wide association study identifies two novel loci and a substantial genetic component for Parkinson's disease. PLoS Genet. 2011;7:e1002141. doi: 10.1371/journal.pgen.1002141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li Y, Zheng H, Luo R, Wu H, Zhu H, Li R, Cao H, Wu B, Huang S, Shao H, et al. Structural variation in two human genomes mapped at single-nucleotide resolution by whole genome de novo assembly. Nat. Biotech. 2011;29:723–730. doi: 10.1038/nbt.1904. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The information in SNPedia is available under a Creative Commons Attribution-Noncommercial-Share Alike 3.0 United States License and also by commercial license. In addition to its wiki web browser interface SNPedia is also available in Distributed Annotation System (DAS) (5) service form from The European Bioinformatics Institute http://www.ebi.ac.uk/das-srv/easydas/bernat/das/SNPedia or in GFF3 at http://www.snpedia.com/files/gbrowse/SNPedia.gff for use with GBrowse (6). The server also understands a Mediawiki API which answers requests in common formats (JSON, XML) and can be accessed with popular libraries in Perl, Python, Java and other languages.