Abstract
The integrated microbial genomes and metagenomes (IMG/M) system provides support for comparative analysis of microbial community aggregate genomes (metagenomes) in a comprehensive integrated context. IMG/M integrates metagenome data sets with isolate microbial genomes from the IMG system. IMG/M's data content and analytical capabilities have been extended through regular updates since its first release in 2007. IMG/M is available at http://img.jgi.doe.gov/m. A companion IMG/M systems provide support for annotation and expert review of unpublished metagenomic data sets (IMG/M ER: http://img.jgi.doe.gov/mer).
INTRODUCTION
The number of metagenome sequence data sets generated by various sequencing centers is rapidly increasing with thousands of data sets already generated. Meteganome sequencing has evolved over the past several years from first generation Sanger (e.g. Applied Biosystems) platforms to second generation 454 Life Sciences Roche (e.g. GS FLX) and Illumina (e.g. GA II and HiSeq) platforms. While cheaper and faster, the new platforms produce shorter sequence fragments (reads). Short read size, higher complexity and inherent incompleteness, make metagenome sequences difficult to assemble and annotate (1,2).
Assembled or unassembled metagenome data sets generated using 454 or Illumina platforms are processed by the IMG/M annotation pipeline (3) before inclusion into IMG/M. Unassembled reads undergo an additional quality control step that includes quality trimming, low-complexity region detection and masking as well as removal of technical replicates. Subsequently, both assembled and unassembled sequences are annotated by the same pipeline that detects CRISPR repeats (4), non-coding RNAs and protein-coding genes (CDSs (Coding Sequence)). RNAs are predicted using tRNAscan-SE (5) for tRNAs, and in-house developed HMM models for rRNAs (6,7,8), while the CDSs are identified using a combination of ab initio gene prediction tools: Prodigal (9), Metagene (10), MetaGenemark (11) and FragGeneScan (12). In addition, sequences in the range of 100–800 bp are compared to the IMG non-redundant protein database using BlastX in order to detect the CDSs missed by ab initio tools. Conflicting gene predictions are consolidated using a weighted schema based on the performance of each method on simulated data sets, with one final gene model generated for each region.
Analysis of the aggregate genomes (metagenomes) of microbial communities (microbiomes) considers the questions of phylogenetic composition and functional or metabolic potential within individual microbiomes, as well as comparisons across microbiomes. IMG/M provides support for such analysis by integrating metagenome data sets with isolate microbial genomes from the integrated microbial genome (IMG) system (13). Using NCBI’s RefSeq (14) as its main source of sequence data, IMG integrates draft and complete microbial genomes from all three domains of life with a large number of plasmids and viruses. Similar to IMG, IMG/M records the primary sequence information for isolate genomes and metagenomes, their organization in scaffolds and/or contigs as well as computationally predicted protein-coding sequences and RNA-coding genes. Protein-coding genes are characterized in terms of additional annotations, such as conserved motifs and domains (15), signal peptides, transmembrane helices (16), pathways and orthology relationships, which may serve as an indication of their functions. These annotations are based on diverse data sources, such as Clusters of Orthologous Genes (COG) clusters and functional categories (17), Pfam (18), TIGRfam and TIGR role categories (19), InterPro domains (20) and KEGG (Kyoto Encyclopedia of Genes and Genomes) Ortholog terms and pathways (21).
We review below IMG/M's data content growth and analysis tool extensions since the last published report on IMG/M (22).
DATA CONTENT
Reference genome data
IMG is the source of IMG/M's reference isolate genomes. The current version of IMG/M is based on the content of IMG 3.4 (V.M. Markowitz et al., submitted publication) consisting of 6891 bacterial, archaeal, eukaryotic and viral genomes, as well as 1186 plasmids that did not come from a specific microbial genome sequencing project, with over 11.6 million protein coding genes.
Genomes generated as part of the Human Microbiome Project (HMP) and the Genome Encyclopedia of Bacterial and Archaea Genomes (GEBA) are of particular importance to metagenome analysis. HMP has generated over 800 reference genomes from both cultured and uncultured bacteria with the goal of supporting the characterization of microbial communities found at multiple human body sites (23). The GEBA project aims at systematically filling the sequencing gaps along the bacterial and archaeal branches of the tree of life (24), with the number of sequenced GEBA genomes standing at 205 as of August 2011. While HMP reference genomes are included into IMG/M from RefSeq via IMG, GEBA genomes are included directly into IMG/M as soon as their annotation is completed at Joint Genome Institute (JGI), before their release through GenBank and RefSeq.
Metagenome data
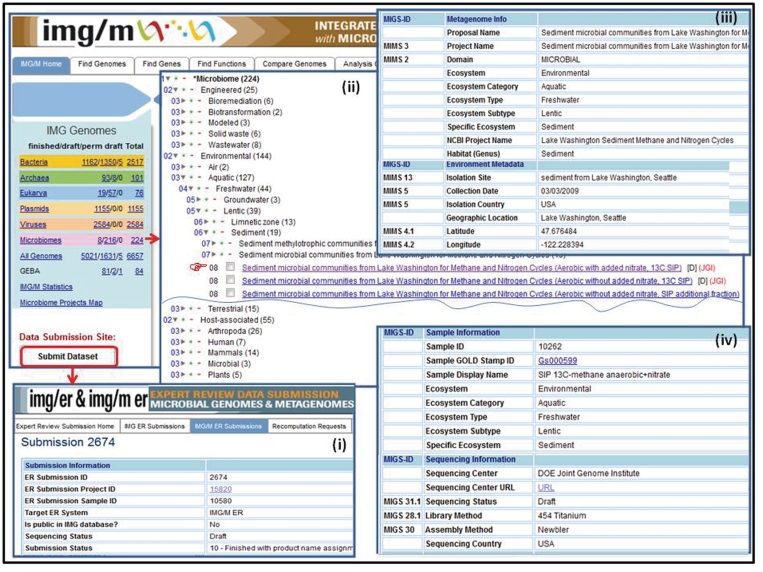
Unlike isolate genomes which are included into IMG and then IMG/M from a public sequence data resource (RefSeq), metagenome data sets are first included into IMG/M ‘Expert Review’ version, IMG/M ER, which allows scientists to employ IMG/M's annotation pipeline as well as review and curate the functional annotation of metagenomes prior to their public release in the context of IMG/M's reference genomes and public metagenomes. Genome and metagenome submissions are handled by the IMG/ER and IMG/M ER submission site, as illustrated in Figure 1(i).
Figure 1.
Metagenome data set classification and metadata characterization. (i) Metagenome data sets are submitted for annotation and inclusion into IMG/M ER via the IMG/ER and IMG/M ER submission site. (ii) Metagenome data sets in IMG/M are organized using a hierarchical classification similar to the phylogenetic classification of isolate genomes. (iii) Metagenome data sets submitted for inclusion into IMG/M ER are associated with metadata characterizing the metagenome study, the associated metagenome sequencing project, environmental information, as well as (iv) sample and sequencing information.
First, the names and classification of metagenome data sets submitted for inclusion into IMG/M ER are curated in GOLD (25) following the five-tiered system as previously proposed (26). This classification scheme underlies the organization of metagenome data sets in IMG/M, as illustrated in Figure 1(ii). Similar to the phylogenetic classification of isolate genomes, the classification of metagenomes is a critical element for conducting metagenome comparative analysis in a rapidly growing universe of metagenome data sets. Thus, all metagenome data sets are organized in three main ecosystem classes: environmental, host associated and engineered classes, then further divided in subclasses characterized by ecosystem categories (e.g. aquatic, terrestrial, air for environmental metagenomes), ecosystem type (e.g. freshwater, marine), ecosystem subtype (e.g. groundwater, drinking water), and specific ecosystem (e.g. cave water, filtered water). Second, metagenome data sets submitted for inclusion into IMG/M ER are associated with comprehensive metadata attributes following the Genome Standards Consortium guidelines (27), as illustrated in Figure 1(iii) and 1(iv). Note that enforcing metadata characterization before metagenome data sets are processed is the most effective way to capture such information.
As of 3 October 2011, IMG/M ER contains about 870 metagenome data sets (samples) with over 163 million protein coding genes that are part of 27 engineered, 110 environmental and 90 host-associated metagenome studies. IMG/M contains the publicly available subset of IMG/M ER metagenome data sets consisting of 289 metagenome data sets with over 60 million protein coding genes, a 10-fold increase compared to August 2007 (22). These data sets are part of 14 engineered, 37 environmental and 32 host-associated studies.
An HMP-specific version of IMG/M, contains 748 metagenome data sets generated as part of the HMP initiative by sequencing samples collected from various body sites (airways, gastrointestinal, oral, skin and urogenital), with a total of 80 million protein-coding genes (http://www.hmpdacc-resources.org/cgi-bin/imgm_hmp/).
DATA ANALYSIS
We briefly review below the IMG/M data analysis tools with emphasis on the support for new metagenome analysis tools developed since the last published report on IMG/M (22).
Data selection and exploration
Metagenomes, genomes, genes and functions can be selected in IMG/M using IMG specific browsers and search tools (15), with the organization of metagenomes using the hierarchical classification discussed above and illustrated in Figure 1 being specific to IMG/M. Metagenomes and genomes that result from search operations are displayed as lists from which they can be selected for inclusion into the ‘Genome Cart’. Genes and functions can be handled in a similar manner using the ‘Gene Cart’ and ‘Function Cart’, respectively.
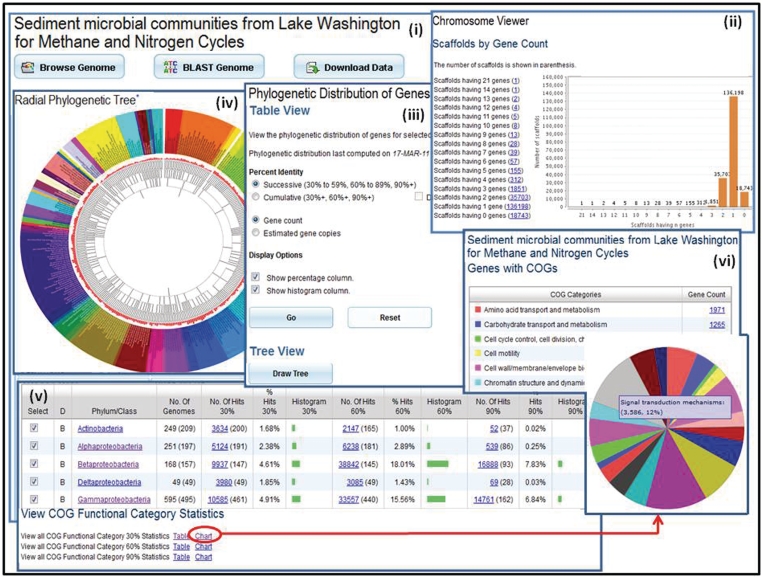
Individual metagenomes can be explored using the ‘Metagenome Details’ page that provides a variety of tools for browsing, searching for the presence of specific genes or downloading metagenome data sets, as illustrated in Figure 2(i). This page also provides information (metadata) on the metagenome together with various statistics of interest, such as the number of genes that are associated with KEGG, COG, Pfam, InterPro or enzyme information.
Figure 2.
Metagenome data exploration. (i) Microbiome samples, such as the Sediment microbial communities from Lake Washington for Methane and Nitrogen Cycle sample, can be examined using the ‘Microbiome Details’ page, which provide tools for browsing, searching or downloading the metagenome data. (ii) ‘Scaffold Cart’ allows selecting individual scaffolds or groups of scaffolds based on properties such as gene content. (iii) The ‘Phylogenetic Distribution of Genes’ provides an estimate of the phylogenetic composition of a metagenome sample based on the distribution of the best BLAST hits of the protein-coding genes in the sample. The result of ‘Phylogenetic Distribution of Genes’ can be displayed using (iv) the ‘Radial Phylogenetic Tree’ viewer or (v) in a tabular format consisting of a histogram with counts protein-coding genes in the sample, which have best BLASTp hits to proteins of isolate genomes in each phylum or class with >90% identity (right column), 60–90% identity (middle column) and 30–60% identity (left column). (vi) The organization of genes by their assignment to COGs is displayed in a pie chart format.
One of the ‘Browse’ tools provided for metagenomes allows examining scaffolds and contigs, whereas a new ‘Scaffold Cart’ allows selecting individual scaffolds (rather than all the scaffolds/contigs of a meteganome) or groups of scaffolds based on their properties such as gene or GC content, scaffold length, read depth, as illustrated in Figure 2(ii), and thus focus the analysis on subsets of metagenome sequences. ‘Scaffold Cart’ provides tools for including the genes of one or several scaffolds into the ‘Gene Cart’, associating a name with selected scaffolds for further analysis, computing a function profile across selected scaffolds, and for examining the phylogenetic distribution of genes for one or several scaffolds in the cart.
The ‘Phylogenetic Distribution of Genes’, illustrated in Figure 2(iii), provides an estimate of the phylogenetic composition of a metagenome sample based on the distribution of the best BLAST hits of the protein-coding genes in the sample. The result of ‘Phylogenetic Distribution of Genes’ can be displayed using the ‘Radial Phylogenetic Tree’ viewer as illustrated in Figure 2(iv), or in a tabular format consisting of a histogram, as illustrated in Figure 2(v) with counts protein-coding genes in the sample, which have best BLASTp hits to proteins of isolate genomes in each phylum or class with >90% identity (right column), 60–90% identity (middle column) and 30–60% identity (left column). This tabular display can be adjusted by filtering out the phyla/classes with few or no hits, whereby the higher the number of hits and percent identity cutoff, the more likely it is that the sample contains close relatives of the sequenced isolate genomes from this phylum/class. The CDSs with best BLAST hits to a certain taxonomic lineage can be organized by their assignment to COGs, which in turn can be classified according to COG Functional Categories (COG Functional Category) or COG Pathways (COG Pathways). The latter can be displayed in a tabular or pie chart format, as illustrated in Figure 2(vi), thereby linking the functional complement of metagenomic proteins with their likely affiliations to different phyla/classes and indicating possible functional specialization within the community (functional guilds). Gene counts in the various display formats of the results are linked to the corresponding lists of genes, which can then be selected and added to ‘Gene Cart’ or analyzed through their ‘Gene Pages’.
The ‘Radial Phylogenetic Tree’ tool allows the comparison of up to five user-selected metagenomes in terms of their BLAST hits to isolate genomes in a color-coded hierarchical circular tree. The resulting tree image can show the hits at different taxonomic levels. More statistics of hits for each genome can be accessed by hovering the mouse over the nodes of the tree. Finally, the genes in a metagenome sample can be viewed in the context of individual reference isolate genome using the ‘Protein Recruitment Plot’ that displays the BLASTp hits of the metagenome genes against the genes of the reference genome, with the coordinates of the scaffold reference genome and the BLAST percent identities shown on the X- and Y-axis, respectively.
Comparative analysis
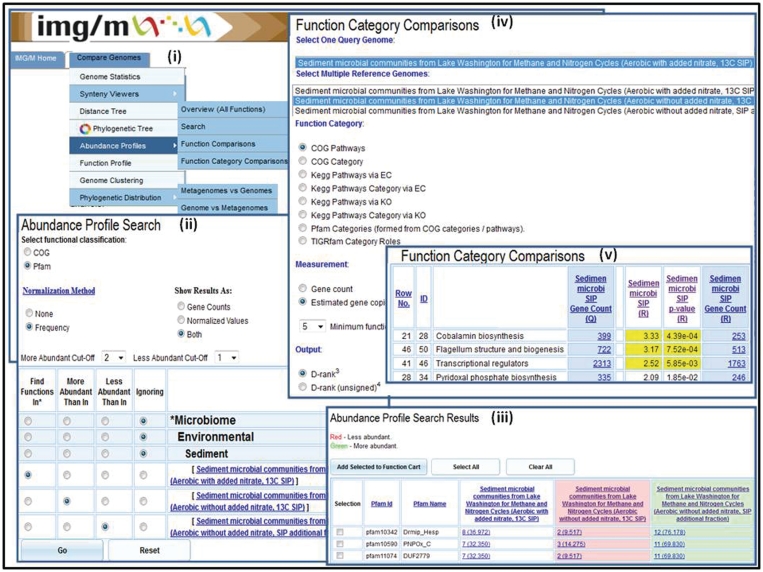
Comparative analysis tools are an extension of the analogous tools in IMG (15), and allow examining the gene content and functional capabilities of microbial communities. We discuss below in more detail the main metagenome-specific comparative analysis tools available under the ‘Compare Genomes’ main menu tab of IMG/M, as shown in Figure 3(i).
Figure 3.
Abundance profile and function comparison tools. The ‘Abundance Profile Search’ allows finding protein families (COGs and Pfams) in metagenomes and isolate genomes based on their relative abundance, such as (ii) finding all Pfams in the Sediment microbial communities from Lake Washington (Aerobic with added nitrate, 13C SIP) sample, which are at least twice as abundant as in the Sediment microbial communities from Lake Washington (Aerobic without added nitrate, 13C SIP) sample and are at least twice less abundant than in Sediment microbial communities from Lake Washington (Aerobic without added nitrate, SIP additional fraction). (iii) The ‘Abundance Profile Search Results’ consists of a list of protein families that satisfy the search criteria together with the metagenomes or genomes involved in the comparison and their associated raw or normalized gene counts. (iv) The ‘Function Category Comparison’ tool allows comparing a metagenome data set with other metagenome data sets or reference genome data sets in terms of the relative abundance of functional categories (COG Pathway, KEGG Pathway, KEGG Pathway Category, Pfam Category and TIGRfam Role Categories). (v) The result of ‘Function Category Comparison’ lists for each function category, F, the number of genes and estimated gene copies in the target (query) metagenome associated with F and for each reference genome/metagenome the number of genes or estimated gene copies associated with F, as well as an assessment of statistical significance in terms of associated P-value and d-rank.
Metagenome samples can be compared in terms of their phylogenetic composition using a variant of the ‘Phylogenetic Distribution of Genes’ tool discussed above, which is extended to allow displaying side by side the phylogenetic distribution of best BLAST hits of protein-coding genes in multiple metagenomes. Two ‘Abundance Profile’ tools allow comparing the functional capabilities of metagenomes and genomes. The ‘Abundance Profile Overview’ tool provides a quick estimate of the functional capabilities of metagenomes in terms of the relative abundance of protein families (COGs and Pfams) and functional families (Enzymes) across selected metagenomes and isolate genomes. The result of this comparison is displayed either as a heat map or in a matrix format, with each column on the map/matrix corresponding to a genome or metagenome, and each row corresponding to a family. Users can ‘drill down’ by following links to lists of genes assigned to a particular family in a specific genome or metagenome.
A new ‘Abundance Profile Search’ tool allows finding protein families (COGs and Pfams) in metagenomes and isolate genomes based on their relative abundance. The tool allows selecting the way the results will be displayed (using raw or normalized gene counts) and setting abundance cutoffs, as illustrated in Figure 3(ii). The ‘Abundance Profile Search Results’ consist of a list of protein families that satisfy the search criteria together with the metagenomes or genomes involved in the comparison and their associated raw or normalized gene counts, as illustrated in Figure 3(iii). Protein families can be selected and added to the ‘Function Cart’, while gene counts are linked to the corresponding lists of genes, which can be subsequently selected and added to the ‘Gene Cart’ for further analysis.
The ‘Abundance Profile’ tools allow comparison of the functional capabilities of metagenomes without assigning statistical significance to the results. However, when metagenomes are compared to each other or to isolate genomes, statistical tests are needed for estimating the statistical significance of the observed differences. The ‘Function Comparison’ and ‘Function Category Comparison’ tools take into account the stochastic nature of metagenome data sets and test whether the differences in abundance can be ascribed to chance variation or not. These tools allow comparing a metagenome data set with other metagenome data sets or reference genome data sets in terms of the relative abundance of (i) protein families (COGs, Pfams and TIGRfams) and functional families (Enzymes) in the case of ‘Function Comparison’ or (ii) functional categories (COG Pathway, KEGG Pathway, KEGG Pathway Category, Pfam Category and TIGRfam subroles) in the case of ‘Function Category Comparison’, as illustrated in Figure 3(iv). The result of these comparisons lists for each function or function category, F, the number of genes or estimated gene copies in the target (query) metagenome associated with F and for each reference genome/metagenome the number of genes or estimated gene copies associated with F. These results include an assessment of statistical significance in terms of associated P-value and d-scores (for Function Comparison) or d-ranks (for Function Category Comparison), as illustrated in Figure 3(v).
FUTURE PLANS
The current version of IMG/M (August 2011) contains 224 metagenome data sets (samples) that are part of 15 engineered, 36 environmental, and 34 host-associated projects (studies). These data sets can be analyzed in the context of 6891 bacterial, archaeal, eukaryotic and virus reference genomes. New metagenome data sets are continuously included into IMG/M from metagenome studies conducted at JGI and other institutes, while new reference isolate genomes are included from IMG on a regular basis.
Data sets from next generation sequencing technology platforms often result in million sequences rendering storing and accessing of data in the standard relational data bases inefficient. As we expect an exponential growth of the size of metagenome data sets by these platforms, we are devising new data management techniques for organizing metagenome data in support of effective analysis.
FUNDING
Director, Office of Science, Office of Biological and Environmental Research, Life Sciences Division, US Department of Energy (Contract No. DE-AC02-05CH11231); National Energy Research Scientific Computing Center, Office of Science of the US Department of Energy (Contract No. DE-AC02-05CH11231); US National Institutes of Health Data Analysis and Coordination Center (Contract U01-HG004866). Funding for open access charge: University of California.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank Shane Cannon of Lawrence Berkeley National Lab's National Energy Research Scientific Computing Center for his help in carrying out large-scale gene similarity computations for IMG/M. We thank Peter Williams, Henrik Nordberg, Roman Nikitin and Simon Minovitsky for their contribution to the development and maintenance of IMG/M. The work of JGI’s production, cloning, sequencing, assembly, finishing and annotation teams is an essential prerequisite for IMG. Eddy Rubin and James Bristow provided support, advice and encouragement throughout this project.
REFERENCES
- 1.Mavromatis K, Ivanova N, Barry K, Shapiro H, Goltsman E, McHardy AC, Rigoutsos I, Salamov A, Korzeniewski F, Land M, et al. On the fidelity of processing metagenomic sequences using simulated dataset. Nat. Methods. 2007;4:495–500. doi: 10.1038/nmeth1043. [DOI] [PubMed] [Google Scholar]
- 2.Wooley JC, Godzik A, Friedberg I. A primer on metagenomics. PLoS Comput. Biol. 2010;6:e1000667. doi: 10.1371/journal.pcbi.1000667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mavromatis K, Ivanova NN, Anderson I, Huntemann M, Williams P, Chen IA, Szeto E, Markowitz VM, Kyrpides NC. The DOE-JGI Standard Operating Procedure for the Annotations of Metagenomes, Standards in Genomic Sciences. 2009;1:63–67. doi: 10.4056/sigs.632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bland C, Ramsey TL, Sabree F, Lowe M, Brown K, Kyrpides NC, Hugenholtz P. CRISPR Recognition Tool (CRT): a tool for automatic detection of clustered regularly interspaced palindromic repeats. BMC Bioinformatics. 2007;8:209. doi: 10.1186/1471-2105-8-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lowe TM, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lagesen K, Hallin P, Rodland EA, Staerfeldt HH, Rognes T, Ussery DW. RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 2007;35:3100–3108. doi: 10.1093/nar/gkm160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Griffiths-Jones S, Moxon S, Marshall M, Khan-na A, Eddy SR, Bateman A. Rfam: annotating non-coding RNAs in complete genomes. Nucleic Acids Res. 2005;33:D121–D124. doi: 10.1093/nar/gki081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nawrocki EP, Kolbe DL, Eddy SR. Infernal 1.0: inference of RNA alignments. Bioinformatics. 2009;25:1335–1337. doi: 10.1093/bioinformatics/btp157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hyatt D, Che GL, LoCascio PF, Land ML, Larimer FW, Hauser LJ. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics. 2010;11:119. doi: 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Noguchi H, Park J, Takagi T. MetaGene: prokaryotic gene finding from environmental genome shotgun sequences. Nucleic Acids Res. 2006;34:5623–5630. doi: 10.1093/nar/gkl723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhu W, Lomsadze A, Borodovsky M. Ab initio gene identification in metagenomic sequences. Nucleic Acids Res. 2010;38:e132. doi: 10.1093/nar/gkq275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rho M, Tang H, Ye Y. FragGeneScan: predicting genes in short and error-prone reads. Nucleic Acids Res. 2010;38:e191–e191. doi: 10.1093/nar/gkq747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Markowitz VM, Chen IA, Palaniappan K, Chu K, Szeto E, Grechkin Y, Ratner A, Anderson I, Lykidis A, Mavromatis K, et al. The integrated microbial genomes (IMG) system: an expanding comparative analysis system. Nucleic Acids Res. 2010;38:D382–D390. doi: 10.1093/nar/gkp887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pruitt KD, Tatusova T, Klimke W, Maglott DR. NCBI Reference Sequences: current status, policy and new initiatives. Nucleic Acids Res. 2009;37:D32–D36. doi: 10.1093/nar/gkn721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moller S, Croning MDR, Apweiler R. Evaluation of methods for the prediction of membrane spanning regions. Bioinformatics. 2001;17:646–653. doi: 10.1093/bioinformatics/17.7.646. [DOI] [PubMed] [Google Scholar]
- 16.Emanuelsson O, Brunak S, von Heijne G, Nielsen H. Locating proteins in the cell using TargetP, SignalP, and related tools. Nat. Protocols. 2007;2:953–971. doi: 10.1038/nprot.2007.131. [DOI] [PubMed] [Google Scholar]
- 17.Tatusov RL, Fedorova ND, Jackson JD, Jacobs AR, Kiryutin B, Koonin EV, Krylov DM, Mazumder R, Mekhedov SL, Nikolskaya AN, et al. The COG database: an updated version includes eukaryotes. BMC Bioinformatics. 2003;4:41. doi: 10.1186/1471-2105-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Finn RD, Mistry J, Tate J, Coggill P, Heger A, Pollington JE, Gavin OL, Gunesekaran P, Ceric G, Forslund K, et al. The Pfam protein families database. Nucleic Acids Res. 2010;38:D211–D222. doi: 10.1093/nar/gkp985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Selengut JD, Haft DH, Davidsen T, Ganapathy A, Gwinn-Giglio M, Nelson WC, Richter AR, White O. TIGRFAMs and Genome Properties: tools for the assignment of molecular function and biological process in prokaryotic genomes. Nucleic Acids Res. 2007;35:D260–D264. doi: 10.1093/nar/gkl1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hunter S, Apweiler R, Attwood TK, Bairoch A, Bateman A, Binns D, Bork P, Das U, Daughterty L, Duquenne L, et al. InterPro: the integrative protein signature database. Nucleic Acids Res. 2005;37:D211–D215. doi: 10.1093/nar/gkn785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kanehisa M, Goto S, Furumichi M, Tanabe M, Hirakawa M. KEGG for representation and analysis of molecular networks involving diseases and drugs. Nucleic Acids Res. 2010;38:D355–D360. doi: 10.1093/nar/gkp896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Markowitz VM, Ivanova N, Szeto E, Palaniappan K, Chu K, Dalevi D, Chen IA, Grechkin Y, Dubchak I, Anderson I, et al. IMG/M: a data management and analysis system for metagenomes. Nucleic Acids Res. 2008;36:D534–D538. doi: 10.1093/nar/gkm869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.The Human Microbiome Jumpstart Reference Strains Consortium. A catalog of reference genomes from the human microbiome. Science. 2010;328:994–999. doi: 10.1126/science.1183605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu D, Hugenholtz P, Mavromatis K, Pukall R, Dalin E, Ivanova NN, Kunin V, Goodwin L, Wu M, Tindall BJ, et al. A phylogeny-driven genomic encyclopaedia of Bacteria and Archaea. Nature. 2009;462:1056–1060. doi: 10.1038/nature08656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liolios K, Chen IM, Mavromatis K, Tavernarakis N, Hugenholtz P, Markowitz VM, Kyrpides NC. The genomes on line database (GOLD) in 2009: status of genomic and metagenomic projects and their associated metadata. Nucleic Acids Res. 2010;38:D346–D354. doi: 10.1093/nar/gkp848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ivanova N, Tringe SG, Liolios K, Liu WT, Morrison N, Hugenholtz P, Kyrpides NC. A call for standardized classification of metagenome projects, Environmen. Microbiol. 2010;12:1803–1805. doi: 10.1111/j.1462-2920.2010.02270.x. [DOI] [PubMed] [Google Scholar]
- 27.Field D, Garrity G, Gray T, Morrison N, Selengut J, Sterk P, Tatusova T, Thomson N, Allen M, Angiuoli SV, et al. Towards a richer description of our complete collection of genomes and metagenomes: the ‘Minimum Information about a Genome Sequence’ (MIGS) specification. Nat. Biotechnol. 2008;26:541–547. doi: 10.1038/nbt1360. [DOI] [PMC free article] [PubMed] [Google Scholar]