Abstract
BRCA1 and BRCA2 are the two main genes responsible for predisposition to breast and ovarian cancers, as a result of protein-inactivating monoallelic mutations. It remains to be established whether many of the variants identified in these two genes, so-called unclassified/unknown variants (UVs), contribute to the disease phenotype or are simply neutral variants (or polymorphisms). Given the clinical importance of establishing their status, a nationwide effort to annotate these UVs was launched by laboratories belonging to the French GGC consortium (Groupe Génétique et Cancer), leading to the creation of the UMD-BRCA1/BRCA2 databases (http://www.umd.be/BRCA1/ and http://www.umd.be/BRCA2/). These databases have been endorsed by the French National Cancer Institute (INCa) and are designed to collect all variants detected in France, whether causal, neutral or UV. They differ from other BRCA databases in that they contain co-occurrence data for all variants. Using these data, the GGC French consortium has been able to classify certain UVs also contained in other databases. In this article, we report some novel UVs not contained in the BIC database and explore their impact in cancer predisposition based on a structural approach.
INTRODUCTION
BRCA1 and BRCA2 are the two main genes predisposing individuals to breast and/or ovarian cancer. Most monoallelic germline mutations in these genes are point variants or deletions/insertions of a few nucleotides. Large rearrangements (exon loss or duplication, 5′ alterations, etc …) have also been described (1,2). These mutations are situated throughout the entire sequence of the two genes. In France, causal mutations in BRCA1 and BRCA2 genes account for about 15% of hereditary breast and ovarian cancers (HBOC) (3).
The pathogenic nature of variants resulting in a premature termination codon (PTC) has been clearly established for those variants leading to nonsense-mediated decay (NMD) (4–7), i.e. BRCA1 proteins with a PTC located upstream from the last 10 carboxy-terminal codons, and BRCA2 proteins with a PTC located upstream from the last 110 carboxy-terminal codons. Some specific missense variants can generate a PTC via abnormal splicing, while others clearly alter the function of BRCA1, including all those occurring in the RING finger and some located in BRCT domains (8). The pathogenicity of these variants has been inferred from linkage analysis of high-risk families, functional assays, biochemical studies and/or demonstration of abnormal mRNA processing (8–10).
The main difficulty encountered in molecular diagnosis of breast and ovarian cancer predisposition relates to the large number of variants with unclear clinical significance. Variants that do not create a PTC represent a significant proportion of DNA variants and are usually called unclassified variants (UVs). UVs mostly consist of missense and intronic variants and in-frame deletions or insertions. Exhaustive collections of UVs could allow cosegregation studies within pedigrees, and these complementary data could help to define a reliable classification. Whether a given UV contributes to the disease phenotype or is merely a neutral variant (or a neutral polymorphism) has direct clinical implications.
Several approaches can be used to determine pathogenicity, including multidisciplinary analysis of cosegregation within pedigrees and degree of family history of the disease, or by indirect measures, including amino acid conservation, severity of amino acid change and evidence from functional assays, outputs from available bioinformatics tools, tumor pathology (11,12), population genetics and comparisons of allele frequencies (13), lack of co-occurrence with causal mutations (14), RNA and protein function studies (15), evolutionary studies of amino acid conservation (16) and a recently proposed combinatorial approach (17). However, such approaches cannot definitively show whether or not a specific variant is causal.
Mutation databases can help to classify variants by providing a unique source of information. The Breast Cancer Information Core Database (http://research.nhgri.nih.gov/projects/bic/index.shtml: BIC) is a voluntary international collection of variants identified worldwide (18). Other online databases include LOVD (Leiden Open Variation Database)-literature unclassified variant (http://chromium.liacs.nl/LOVD2/cancer/home.php) (19), LOVD-IARC (http://brca.iarc.fr/LOVD/home.php), kConFab (Kathleen Cuningham Foundation Consortium for Research into Familial Breast Cancer) (20), The Singapore Human Mutation and Polymorphism Database (21), and HGMD (The Human Gene Mutation Database at the Institute of Medical Genetics in Cardiff) (22), NGRL/DmuDb (https://ngrl.manchester.ac.uk/Browser/genes.htm).
In this report, we present exhaustive databases of French BRCA1/2 variants (UMD-BRCA1/BRCA2: http://www.umd.be/BRCA1/ and http://www.umd.be/BRCA2/) developed with UMD software (23). These databases have been endorsed by the French Cancer National Institute (INCa) and are available online. A curator collects and compiles information from 16 licensed laboratories belonging to the French GGC consortium (Groupe Génétique et Cancer) and located all over France. UMD-BRCA1/BRCA2 databases centralize all variants (causal, neutral and UVs), allowing identification of co-occurrence data. Data on polymorphisms were not collected. In October 2011, the UMD-BRCA1/BRCA2 databases contained data from a set of 3743 families for BRCA1 and 3,650 families for BRCA2. In this report, we first analyze BRCA1/BRCA2 variants in the French population, and then use our co-occurrence data to classify some UVs as either causal or neutral. Finally, we describe BRCA1 and BRCA2 UVs and evaluate their pathogenicity, notably from a structural standpoint (Supplementary Data). Therefore, this database should be a tool to classify variant and promote specific studies.
MATERIALS AND METHODS
Software and search engine
The UMD-BRCA1/BRCA2 databases were constructed with an updated version of the Universal Mutation Database generic software (http://www.umd.be) (23,24), which includes an optimized structure to assist and secure data entry and to allow the input of various clinical or biological data. An offline copy of UMD-BRCA1/BRCA2 is continuously edited and updated by the curators; edited copies are regularly deposited on the server. These databases are freely available online. Their integrity is ensured by the original (edited) template offline. Notification of errors in the current version would be gratefully received by the curator author. Database design follows general recommendations for LSDB characteristics and their curation as proposed by the Human Genome Variation Society (HGVS) (25).
Website
The Website is divided into six sections, four of which are dedicated to information on the BRCA1 or BRCA2 genes and numbering of exons and cDNA sequences, information on the protein and the references, while two sections allow the user to search into the database. Hyperlinks are provided to access the reference sequences and data search capabilities. The web pages also contain links to external information including central biological databases, such the Human Genome Database (GDB), Pubmed, GeneCard, UniGene, Uniprot, OMIM, HGV, etc.
Probands
About 24 000 probands were managed at the medical genetics departments of French Comprehensive Cancer Care centers and hospitals between January 1995 and October 2011. Eligibility criteria for familial genetic testing are described in the French consensus statement (26). All probands enrolled in this study gave their written informed consent for specific genetic testing. The probands were the index cases of individual families and were screened for BRCA1 and BRCA2 variants. Records in the French UMD-BRCA1/BRCA2 databases are restricted to index cases of families with clearly causal mutations, neutral variants or UVs. A system of five classes has been used for classification (27). An anonymized and unique sample identifier (sample_ID) is generated for each family.
Variant screening
The full coding sequences and exon–intron junctions of the BRCA1 and BRCA2 genes were screened for variants, based on prescreening (DGGE, SSCP, PTA, dHPLC, HRM or EMMA) and sequencing. Several large rearrangements were identified by large cDNA sequencing, MLPA (28), QMPSF (29), qPCR (30), qPCR HRM (31), EMMA (32), bar code screening (33) or dedicated array CGH (34).
UMD-BRCA1/BRCA2 databases
The UMD software includes an automatic procedure to check for the correct description of the sequence variation at the nucleotide level and to generate the variant name at the protein level. All variant types are documented. All variants were subjected to a curation process and named according to the HGVS nomenclature before integration in the databases. Variants previously described either in publications or in other BRCA1/2 databases were reviewed. Data on 7396 different BRCA1/BRCA2 families were collected between 1995 and October 2011 (Table 2). Co-occurrence of each UV with causal mutations was recorded in the databases. Data on polymorphism were not collected. Families were recorded only once, independently of the number of members tested. No clinical data were collected.
Table 2.
Families and records distribution in the UMD-BRCA1/BRCA2 databases
| Causal mutation | UV | 1 causal mutation + x UV | Total of families | |
|---|---|---|---|---|
| BRCA1 5311 entries/3743 families | ||||
| Families with 1 variation | 2636 | 948 | – | 3584 |
| Families with 2 variations | 8 | 50 | 76 | 134 |
| Families with 3 variations | – | 16 | 3 | 19 |
| Families with 4 variations | – | 4 | 2 | 6 |
| Total of families | 2644 | 1018 | 81 | 3743 |
| BRCA2 5532 entries/3650 families | ||||
| Families with 1 variation | 1576 | 1775 | – | 3351 |
| Families with 2 variations | 1 | 141 | 116 | 258 |
| Families with 3 variations | – | 19 | 12 | 31 |
| Families with 4 variations | – | 5 | 1 | 6 |
| Families with 5 variations | – | 2 | – | 2 |
| Families with 6 variations | – | 2 | – | 2 |
| Total of families | 1577 | 1944 | 129 | 3650 |
Causal mutations and UVs. There are three supplementary families with 1 causal BRCA1 mutation and 1 causal BRCA2 mutation.
The present version of BRCA1 and BRCA2 is based on GenBank reference sequences NM_007294.2 (mRNA: U14680) and NM_000059.3 (mRNA: U43746), respectively. For BRCA1, this entry includes 119 bases of 5′-untranslated sequence with protein translation starting at nucleotide 120. The reference BRCA2 sequence also includes the 228-base 5′-untranslated region.
Searching tools
The UMD knowledgebase system includes many analytical tools (23,24). In addition to previously developed routines, new tools have been specifically developed for the UMD-BRCA1/BRCA2 for in-depth analysis. The user is able to optimize multicriteria search tools to select records from any field.
Prequeried data can be displayed by several Searching Tools in ‘Mutations’ such as ‘Type and number of mutations’ or ‘Mutation by exon/intron’ (Supplementary Figure S1A). Alternatively, the option ‘I found a mutation’ provides access to a quick search (nucleotide or amino acid position), ‘Free search’ provides access to an advanced search and a customized search interface searching several items (sample ID, amino acid position, biological significance, etc). A simple click on hyperlinks provides access to a list of interesting variants (Supplementary Data).
Variant analysis
For co-occurrence analysis, a UV in trans with two different causal mutations, was considered to be unlikely to cause disease.
The impact of missense substitutions in the BRCA1/2 genes was predicted by using three online programs: Align-GVGD (35), SIFT (36) and PolyPhen (37), which predict the effect of missense substitutions on gene function, as well as the new UMD-Predictor® tool included in the UMD package (17).
For structural analysis, we focused on some novel variants not contained in the BIC database and located in a conserved region or in a previously reported functional domain. Regions and domains of BRCA1 or BRCA2 are described in Supplementary Data (Supplementary Figure S2).
RESULTS
UMD-BRCA1/BRCA2 databases
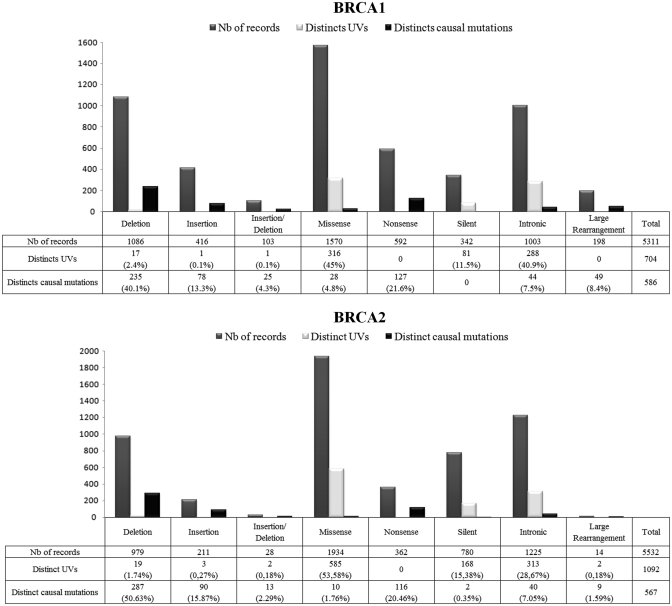
In UMD-BRCA1, a total of 1391 distinct BRCA1 variants comprising 586 causal mutations (42%), 5 likely causal mutations (0.4%), 22 likely neutral variants (1.6%), 74 neutral variants (5%) and 704 UVs (51%) have been identified in 3743 French families (1 index case harbors two different causal BRCA1 mutations and 81 cases harbor one causal mutation associated with at least one UV) (Tables 1 and 2). In UMD-BRCA2, a total of 1704 distinct BRCA2 variants were reported in 3650 families, with 567 different causal mutations (33.1%), 17 likely neutral variants (1%), 30 neutral variants (1.8%) and 1092 UVs (64.1%) (1 index case harbors two different causal BRCA2 mutations and 129 cases harbor one causal mutation and at least one UV) (Tables 1 and 2). Three index cases were double heterozygotes for causal BRCA1 and BRCA2 mutations. Most variants were found in a unique family (517 of 704 BRCA1 UVs and 738 of 1092 BRCA2 UVs) (Table 1). A total of 3743 BRCA1 families (2636 causal mutations and 948 UVs) and 3650 BRCA2 families (1576 causal mutations and 1775 UVs) were found to have at least one mutation (Table 2).
Table 1.
The October 2011 issue of UMD-BRCA1/BRCA2 databases
| Causal | Likely causal | UV | Likely neutral | Neutral | Total | |
|---|---|---|---|---|---|---|
| BRCA1 | ||||||
| Records | 2733 (51.4%) | 17 (0.3%) | 1200 (22.6%) | 190 (3.6%) | 1171 (22.1%) | 5311 |
| Distinct variants | 586 | 5 | 704 | 22 | 74 | 1391 |
| Unique variants | 311 | 1 | 517 | 5 | 9 | 843 |
| BRCA2 | ||||||
| Records | 1707 (30.9%) | – | 2289 (41.4%) | 387 (7%) | 1149 (20.7%) | 5532 |
| Distinct variants | 567 | – | 1092 | 17 | 30 | 1704 |
| Unique variants | 337 | – | 738 | 3 | 0 | 1078 |
Causal mutations, UVs and neutral
The exon distribution of the UVs, neutral variants and causal mutations in the UMD-BRCA1/BRCA2 databases was also assessed and no statistically significant differences were found between the UMD-BRCA1/BRCA2 and BIC databases (see Supplementary Data). Based on the ratio between UMD-BRCA1/BRCA2 variants and predicted variants, BRCA1 exons 2, 3, 5, 17, 18, 19 and 21 are the most frequently mutated exons (Supplementary Table S1A). Exons 2, 3, and 5 correspond to the RING-finger, exons 17, 18 and 19 to the first BRCT domain and exon 21 to the second BRCT. In BRCA2, exons 2, 3, 18 and 23 are the most frequently mutated exons (Supplementary Table S1B). Exon 18 correponds to part of HD and OB1 domains and exon 23 correponds to part of the OB2 domain. Structural data have been previously available for only a small segment of exon 3 (residues 21–39), and no 3D structures have been described or predicted for exon 2.
Characterization of French causal mutations in the UMD-BRCA1/BRCA2 databases
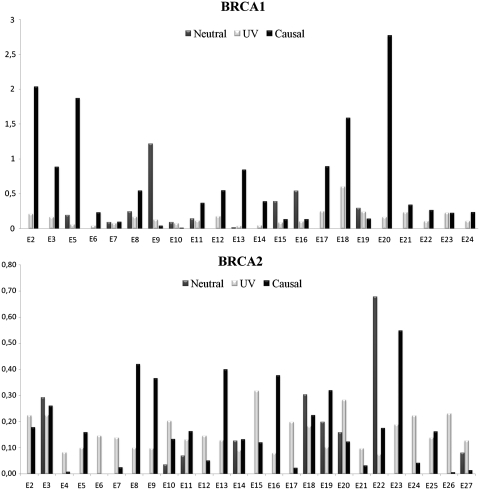
The spectrum of causal mutations in the BRCA1 and BRCA2 genes involves the entire sequence (Figure 1). The BRCA1 exons most frequently mutated are exons 2, 5 and 20 and the BRCA2 exons most frequently mutated are exons 8, 13 and 23 (Figure 1).
Figure 1.
Distribution of all variants in the UMD-BRCA1/BRCA2 databases. The number of entries of all variants is represented per nucleotide and per exon of BRCA1 or BRCA2.
The most prevalent causal BRCA1 mutations identified in the French population were: the French founder frameshift c.3481_3491del (3600del11, exon 11) from North-Eastern France (137 records) (38,39) and c.5266dup (5382insC, exon 20), a founder Ashkenazi Jewish mutation (138 records). This causal mutation was found 48 times in Ile-de-France and 27 times in North-Eastern France. Six other frequent recurrent causal BRCA1 mutations were observed more than 8 times (Table 3): all had been recorded early in the BIC database (October 2011), except for c.5128G>T (9 records), which was found 8 times in North-Eastern France (39).
Table 3.
Geographical distribution of founder causal mutations in BRCA1 and BRCA2 genes
| BRCA1 | ||||||||
|---|---|---|---|---|---|---|---|---|
| c.68_69delAG exon 2 | c.3839_3843delCTCAG exon 11 | c.3481_3491del11 exon 11 | c.3841C>T exon 11 | c.4327C>T exon 13 | c.5030_5033delCTAA exon 17 | c.5128G>T exon 18 | c.5266dup exon 20 | |
| (185delAG) | (3958del5ins4) | (3600del11) | (3960C>T) | (4446C>T) | (5149delCTAA) | (5247G>T) | (5382insC) | |
| Ashkenazi Jews | French | North-Eastern France | France/Belgium/Holland | French Canadian | French | North-Eastern France | Ashkenazi Jews | |
| IDF | 28 | 12 | 18 | 6 | 17 | 13 | – | 48 |
| NW | 1 | 15 | 4 | 2 | 25 | 3 | – | 11 |
| North | 1 | – | 2 | 13 | – | 7 | – | 5 |
| NE | 5 | 3 | 94 | 4 | 1 | 1 | 8 | 27 |
| SE | 14 | – | 12 | – | 1 | 5 | – | – |
| SW | 1 | – | 3 | 4 | 2 | 3 | – | 11 |
| Center | 5 | 11 | 3 | 2 | 2 | – | 1 | 14 |
| UMD database (3158 records) | 56 (1.77%) | 41 (1.29%) | 137 (4.34%) | 31 (0.98%) | 48 (1.52%) | 32 (1.01%) | 9 (0.29%) | 138 (4.37%) |
| BIC database (11329 records) | 1980 (17.5%) | 3 (0.03%) | 58 (0.51%) | 12 (0.11%) | 126 (1.11%) | 17 (0.15%) | 0 | 1063 (9.38%) |
| BRCA2 | ||||||||
|---|---|---|---|---|---|---|---|---|
| c.771_775delTCAAA exon 9 | c.1310_1313delAAGA exon 10 | c.2808_2811delACAA exon 11 | c.5645C>A exon 11 | c.5946delT exon 11 | c.6275_6276delTT exon 11 | c.6644_6647delACTC exon 11 | c.9026_9030delATCAT exon 23 | |
| (999del5) | (1538del4) | (3036del4) | (5873C>A) | (6174delT) | (6503delTT) | (6872del4) | (9254del5) | |
| Iceland | European | Western Europe | East European | Ashkenazi Jews | Dutch | French | Northeast Spanish | |
| IDF | 4 | 8 | 21 | 8 | 13 | 8 | 4 | 14 |
| NW | – | 1 | 5 | – | 1 | 2 | 1 | – |
| North | – | – | 1 | – | – | 1 | 1 | – |
| NE | – | 3 | 6 | 7 | 2 | – | 5 | 1 |
| SE | 5 | 5 | 3 | 1 | 2 | 3 | 3 | 7 |
| SW | – | – | 3 | 1 | – | – | 3 | 3 |
| Center | 1 | 3 | 2 | – | 1 | 1 | 1 | – |
| UMD database (3134 records) | 10 (0.32%) | 20 (0.64%) | 41 (1.3%) | 17 (0.54%) | 19 (0.6%) | 15 (0.48%) | 18 (0.57%) | 25 (0.80%) |
| BIC database (11 330 records) | 14 (0.12%) | 10 (0.09%) | 105 (0.93%) | 26 (0.23%) | 1087 (9.49%) | 93 (0.82%) | 20 (0.18%) | 10 (0.09%) |
Numbers of families with founder mutations throughout the country (Polymorphisms are excluded); IDF: Paris; NW: Caen, Nantes; North: Lille; NE: Nancy, Reims, Strasbourg; SE: Montpellier, Marseille, Nice; SW: Bordeaux, Toulouse; Center: Lyon, Clermont-Ferrand. *χ2 test.
We identified eight frequent causal BRCA2 mutations (Table 3), all of which were contained in the BIC database (August 2011). The most prevalent causal BRCA2 mutation in the French population is c.2808_2811delACAA, a Western European mutation (41 records) (40). All of these eight recurrent causal BRCA2 mutations were frequently detected in the Paris region.
In comparison with the BIC database, the UMD database also identified causal mutations from the Ashkenazi Jewish population (26.88% for BIC-BRCA1 versus 6.14% UMD-BRCA1, 9.49% for BIC-BRCA2 versus 0.6% for UMD-BRCA2), but also several frequent causal mutations from other European ethnic populations.
Characterization of French UVs in the UMD-BRCA1/BRCA2 databases
UVs were also detected throughout the two genes. The most frequently mutated exons are 17, 18 and 19 for BRCA1 and 15, 20 and 26 for BRCA2 (Figure 1).
As expected, numerous variants (at least 51% of distinct variants in BRCA1 and 64.1% of distinct variants in BRCA2) could not be classified as causal or neutral. These UVs were found in BRCA1 in 1018 families and in BRCA2 in 1944 families. In BRCA1, 704 distinct UVs were identified in one or several families: 948 families with one BRCA1 UV only, 81 with a UV associated with a causal mutation and 70 with UVs only (Tables 1 and 2). In BRCA2, 1092 distinct UVs were identified: 1775 families with one BRCA2 UV only, 129 with a UV associated with a causal mutation and 169 with UVs only (Tables 1 and 2).
Most UVs were observed only once (517/704 in BRCA1 and 738/1092 in BRCA2) (Table 1), while some were recurrent, including BRCA1 c.5075-53C>T (IVS17-53C>T, SNP rs8176258) with 66 entries, c.5277+48_59dup (IVS20+48_59dup, 47 entries) and BRCA2 c.7008-62A>G (IVS13-62A>G, 69 entries), c.9257-16T>C (IVS24-16T>C, SNP rs11571818, 50 entries) and c.6100C>T (p.Arg2034Cys, exon 11, SNP rs1799954, 85 entries).
Most UVs identified in this study were either missense changes (45% in BRCA1 and 53.58% in BRCA2) or silent variants (11.5% in BRCA1 and 15.38% in BRCA2) (Figure 2). Small deletions or insertions of various lengths were also observed (2.6% in BRCA1 and 2.19% in BRCA2) (Figures 2), as well as intronic variants (40.9% in BRCA1 and 15.38% in BRCA2) (Figure 2). Intronic variants lie outside intron–exon boundaries and their potential impact on the splicing process is difficult to predict and need specific RNA analysis (Figure 2).
Figure 2.
Summary of all allelic variants contained in the UMD-BRCA1/2 mutation databases. Total number of types of variants in the BRCA1 and BRCA2 genes.
GGC classification of some UVs already reported in other databases
Co-occurrence data on a UV in trans of a known causal mutation (the UV and the causal mutation are on opposite chromosomes) can provide a powerful means of ruling out pathogenicity (14). Identification of co-occurrence of a UV and a causal mutation requires further investigations such as cloning to exclude the in cis position of these two variants (41). Without these additional analyses, there is still a very weak probability that the two causal mutations are situated in cis, which is why the following hypothesis can be used: if a UV coexists with at least two different causal mutations in two different families, this UV can be considered to be always in trans with respect to the causal mutation. To support the co-occurrence results, we also used an integrated approach combining the Align-GVGD, SIFT and PolyPhen programs, published data, and the UMD-Predictor tool, allowing 3 BRCA2 UVs to be classified as neutral and polymorphism (Table 4). These BRCA2 UVs, described in the BIC database, were reported in our databases as co-occurring with causal mutations: c.6100C>T [p.Arg2034Cys, exon 11, 137 records, 16 in trans (9 distinct causal mutations)], c.7008-62A>G (IVS13-62A>G, 106 records, 6 distinct causal mutations in trans), and c.9257-16T>C [IVS24-16T>C, 55 records, 8 in trans (4 distinct causal mutations)]. This suggests that these variants do not sufficiently alter BRCA2 function to predispose to cancer. The status of the BRCA2 c.6100C>T variant differed between the algorithms: it was predicted to be pathogenic by the UMD-Predictor® tool and neutral by SIFT, Align-GVGD, PolyPhen and co-occurrence analysis. Moreover, c.6100C>T has been described as a polymorphism in a Spanish population of cancer families and patients (42). We classified this variant as a neutral variant. Our classification is reported in the UMD-BRCA1/BRCA2 databases. When it possible, a structral approach is used to help classify the UVs (see example in Supplementary Data: A structural approach to novel UVs).
Table 4.
Variants classified by GGC
| BRCA2 |
|||
|---|---|---|---|
| Variant (cDNA level) | c.6100C>T | c.7008-62A>G | c.9257-16T>C |
| Variant (protein level) | p.Arg2034Cys | – | – |
| Reported in UMD-BRCA1/BRCA2 | 137 | 106 | 55 |
| Co-occurrence with a causal mutation | 16 (9) | 6 | 8 (4) |
| dbSNP | Rs1 799 954 | – | rs11 571 818 |
| Align-GVGD | GV = 257.44, GD = 11.33, C0 (less likely) | ND | ND |
| PolyPhen | 0.423 (benign) | ND | ND |
| SIFT | 0.07 (no deleterious) | ND | ND |
| UMD-Predictor® | 82 (pathogenic) | ND | ND |
| Literature data | Polymorphism | Neutral | Neutral/Polymorphism |
| Classification BIC database | UV (97) | UV (2) | UV (6) |
| Classification UMD-BRCA1/BRCA2 database | Polymorphism | Neutral | Neutral |
The enriched deleterious group consisted of those substitutions with Align-GVGD scores of GV<62 and GD>0 and SIFT scores ≤0.05. The enriched neutral group consisted of those substitutions with Align-GVGD scores of GV>0 and GD <62 and SIFT scores >0.1. PolyPhen results for each variant were classified as benign (score ≤ 0.5), possibly damaging (0.5< score< 2), probably damaging (score > 2), and unknown. UMD predictor® results for each missense were classified as polymorphism (score ≤ 50), probable polymorphism (50 ≤ score ≤ 64), probably pathogenic (65 ≤ score ≤ 74) and pathogenic (score > 74). ND: not determined because algorithms only predict the impact of missense mutations.
DISCUSSION
The UMD-BRCA1/2 databases provide an overview of the BRCA1/2 variants in the French population
We examined BRCA1/2 UVs collected by 16 licensed laboratories located throughout France and belonging to the French GGC consortium ‘Groupe Génétique et Cancer’, which is designed to validate diagnostic procedures and strategies. The 16 licensed laboratories automatically send all results of families tested in France with a BRCA1/2 variant. Every 3 months, an update is performed by all laboratories. The UMD-BRCA1/BRCA2 databases have been approved by the French National Cancer Institute (INCa).
Among 24 000 families tested, 2733 BRCA1 and 1707 BRCA2 families with one causal mutation were identified, as well as 1018 BRCA1 and 1,944 BRCA2 families with at least one UV (Table 2). About 95% of BRCA1/2 variants (causal mutations and UVs) were observed only once.
Certain exons showed an excess of variants. Interestingly, in BRCA1, these exons correspond to the RING-finger domain and the first BRCT domain, while in BRCA2, the most frequently mutated exons are not associated with resolved structural domains (Supplementary Tables S1A and S1B).
As in other populations, some variants are recurrent in the French population (40,43–45). Sixteen recurrent causal mutations in BRCA1 and BRCA2 were reported more than eight times each. Some UVs were also recurrent. Three were most frequent in BRCA2, namely c.7008-62A>G (106 families), c.9257-16T>C (55 families) and c.6100C>T (137 families). These three variants could be geographic polymorphisms, or specific to the French population. Detection of c.7008-62A>G and c.9257-16T>C could also be variable according to the positions of the primers used to amplify the region and the frequency differs from one database to another due to the selection of PCR primers.
The UMD-BRCA1/BRCA2 databases and pooled data from the French GGC are sufficiently powerful for co-occurrence studies. If homozygous and compound heterozygous variants in BRCA1 are assumed to be embryonically lethal (46–48), while those in BRCA2 are responsible for Fanconi anemia (49–51), several variants can be classified as neutral based on a single observation. One patient reported in the UMD-BRCA1/BRCA2 databases harbors two different causal BRCA1 mutations in cis (Bezieau, S. and Delnatte, C. unpublished data). Another patient harbors two different causal BRCA2 mutations. These two causal mutations result in a breast/ovarian cancer syndrome but also in glioblastoma (Sevenet, N., Longy, M. et al. unpublished data).
A double heterozygote with a causal BRCA1 mutation and a causal BRCA2 mutation did not appear to present a more severe phenotype (52). We also identified three families that included double heterozygotes with a causal mutation in BRCA1 and a causal mutation in BRCA2. Coexistence of causal mutations in BRCA1 and BRCA2 is unusual in non-Ashkenazi populations. Two of these three families did not harbor the Ashkenazi founder mutations (unpublished data) (52), but these three families presented a breast cancer and other cancers syndrome.
UVs classified by GGC
Most information used to determine the pathogenicity of an UV is indirect, such as segregation data, loss of heterozygosity, absence of the UV in an unaffected subject or in silico prediction based on the conservation, position and nature of amino acid change or the impact on the mRNA.
We used co-occurrence data and predictive algorithms to classify three BRCA2 variants (Table 4). The French consortium uses these data to classify UVs in the UMD-BRCA1/BRCA2 databases. For example, like Goldgar et al., we classified the BRCA2 c.6100C>T (p.Arg2034Cys) variant as a neutral variant (53). In our database, this c.6100C>T variant was associated with eight distinct causal mutations strongly suggesting that it is not causal. However, the status of the BRCA2 c.6100C>T variant differed between the algorithms: SIFT score was 0.07, corresponding to borderline pathogenicity (0.05). The UMD Predictor® tool contained the score SIFT but also other scores. The final conclusion of neutrality was based on a combination of prediction programs with co-occurrence data. We discuss the contribution of the 3D structure in Supplementary Data.
CONCLUSION
BRCA1 and BRCA2 are the two genes principally involved in predisposition to breast and ovarian cancer. Classification of UVs of BRCA1 and BRCA2 has major implications for hereditary forms of these malignancies. Most variants identified to date cannot yet be definitively classified as causal or neutral. The French UMD-BRCA1/BRCA2 databases, which collect all variants (neutral, causal and UVs), differ from other BRCA databases by the fact that they contain co-occurrence data for all variants, allowing the GGC consortium to classify a number of UVs. In the future, co-occurrence data and structural approaches should help to classify further unclassified BRCA1/BRCA2 variants.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1 and 2, Supplementary Figures 1–7, Supplementary References [8,17,23,27,54–61].
FUNDING
French National Cancer Institute (INCa) (also to S.C. and L.B.); Development of the UMD tool used to construct the UMD-BRCA1/BRCA2 databases received funding from the European Community's Seventh Framework Program (FP7/2007-2013) under grant agreement no. 200754—the GEN2PHEN project; Association d'Aide à la Recherche Cancérologique de Saint Cloud (ARCs). Funding for open access charge: Institut Curie—Hospital René Huguenin.
Conflict of interest statement. None declared.
ACKNOWLEDGMENTS
The authors thank the French oncogeneticists, the GGC leads by Dr Catherine Noguès and probands for their cooperation.
APPENDIX
French BRCA GGC consortium: Nicolas Sevenet, Françoise Bonnet, Michel Longy: Institut Bergonié—Bordeaux; Agnès Hardouin, Dominique Vaur, Sophie Krieger: Centre François Baclesse—Caen; Nancy Uhrhammer, Yves-Jean Bignon: Centre Jean Perrin—Clermont-Ferrand; Jean-Philippe Peyrat, Françoise Revillion, Joëlle Fournier: Centre Oscar Lambret—Lille; Audrey Remenieras, Sylvie Mazoyer: Centre de Recherche en Cancérologie de Lyon—Lyon; Mélanie Léone: Hospices Civils de Lyon and Centre Léon Bérard—Lyon; Hagay Sobol, Tetsuro Noguchi, Violaine Bourdon, Audrey Remenieras: Institut Paoli-Calmettes—Marseille; Jean-Marc Rey: Laboratoire de Biologie Cellulaire et Hormonale (CHU Arnaud de Villeneuve)—Montpellier; Myriam Bronner, Philippe Jonveaux, Christophe Philippe: CHU de Nancy-Brabois—Vandœuvre-lès-Nancy; Capucine Delnatte: CHU—Institut de Biologie—Hôtel Dieu—Nantes; Florence Coulet—Groupe hospitalier Pitié-Salpêtrière, Assistance Publique-Hôpitaux de Paris, Université Pierre et Marie Curie, Laboratoire d'Oncogénétique et Angiogénétique moléculaire—Paris; Laurent Castera, Virginie Caux-Moncoutier, Claude Houdayer, Dominique Stoppa-Lyonnet: Institut Curie—Paris; Chantal Delvincourt, Marie-Claude Gorisse: CHU et Institut Jean Godinot—Reims; Ivan Bièche, Cédrick Lefol: Institut Curie—Hôpital René Huguenin, Service d'Oncogénétique—Saint Cloud; Danièle Muller, Joseph Abecassis: Centre Paul Strauss—Strasbourg; Christine Toulas: Institut Claudius Régaud—Toulouse; Marine Guillaud-Bataille, Brigitte Bressac-De Paillerets: Institut Gustave Roussy—Villejuif.
REFERENCES
- 1.Mazoyer S. Genomic rearrangements in the BRCA1 and BRCA2 genes. Hum. Mutat. 2005;25:415–422. doi: 10.1002/humu.20169. [DOI] [PubMed] [Google Scholar]
- 2.Sluiter MD, van Rensburg EJ. Large genomic rearrangements of the BRCA1 and BRCA2 genes: review of the literature and report of a novel BRCA1 mutation. Breast Cancer Res. Treat. 2010;125:325–349. doi: 10.1007/s10549-010-0817-z. [DOI] [PubMed] [Google Scholar]
- 3.INCa. Consultations et Laboratoires. Boulogne-Billancourt: Institut National du Cancer; 2010. [Google Scholar]
- 4.Ware MD, DeSilva D, Sinilnikova OM, Stoppa-Lyonnet D, Tavtigian SV, Mazoyer S. Does nonsense-mediated mRNA decay explain the ovarian cancer cluster region of the BRCA2 gene? Oncogene. 2006;25:323–328. doi: 10.1038/sj.onc.1209033. [DOI] [PubMed] [Google Scholar]
- 5.Anczukow O, Ware MD, Buisson M, Zetoune AB, Stoppa-Lyonnet D, Sinilnikova OM, Mazoyer S. Does the nonsense-mediated mRNA decay mechanism prevent the synthesis of truncated BRCA1, CHK2, and p53 proteins? Hum. Mutat. 2008;29:65–73. doi: 10.1002/humu.20590. [DOI] [PubMed] [Google Scholar]
- 6.Caux-Moncoutier V, Pages-Berhouet S, Michaux D, Asselain B, Castera L, De Pauw A, Buecher B, Gauthier-Villars M, Stoppa-Lyonnet D, Houdayer C. Impact of BRCA1 and BRCA2 variants on splicing: clues from an allelic imbalance study. Eur J Hum Genet. 2009;17:1471–1480. doi: 10.1038/ejhg.2009.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen X, Truong TT, Weaver J, Bove BA, Cattie K, Armstrong BA, Daly MB, Godwin AK. Intronic alterations in BRCA1 and BRCA2: effect on mRNA splicing fidelity and expression. Hum. Mutat. 2006;27:427–435. doi: 10.1002/humu.20319. [DOI] [PubMed] [Google Scholar]
- 8.Carvalho MA, Marsillac SM, Karchin R, Manoukian S, Grist S, Swaby RF, Urmenyi TP, Rondinelli E, Silva R, Gayol L, et al. Determination of cancer risk associated with germ line BRCA1 missense variants by functional analysis. Cancer Res. 2007;67:1494–1501. doi: 10.1158/0008-5472.CAN-06-3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sinilnikova OM, Mazoyer S, Bonnardel C, Lynch HT, Narod SA, Lenoir GM. BRCA1 and BRCA2 mutations in breast and ovarian cancer syndrome: reflection on the Creighton University historical series of high risk families. Fam. Cancer. 2006;5:15–20. doi: 10.1007/s10689-005-2571-7. [DOI] [PubMed] [Google Scholar]
- 10.Morris JR, Pangon L, Boutell C, Katagiri T, Keep NH, Solomon E. Genetic analysis of BRCA1 ubiquitin ligase activity and its relationship to breast cancer susceptibility. Hum. Mol. Genet. 2006;15:599–606. doi: 10.1093/hmg/ddi476. [DOI] [PubMed] [Google Scholar]
- 11.Goldgar DE, Easton DF, Byrnes GB, Spurdle AB, Iversen ES, Greenblatt MS. Genetic evidence and integration of various data sources for classifying uncertain variants into a single model. Hum. Mutat. 2008;29:1265–1272. doi: 10.1002/humu.20897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goldgar DE, Easton DF, Deffenbaugh AM, Monteiro AN, Tavtigian SV, Couch FJ. Integrated evaluation of DNA sequence variants of unknown clinical significance: application to BRCA1 and BRCA2. Am. J. Hum. Genet. 2004;75:535–544. doi: 10.1086/424388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee E, McKean-Cowdin R, Ma H, Chen Z, Van Den Berg D, Henderson BE, Bernstein L, Ursin G. Evaluation of unclassified variants in the breast cancer susceptibility genes BRCA1 and BRCA2 using five methods: results from a population-based study of young breast cancer patients. Breast Cancer Res. 2008;10:R19. doi: 10.1186/bcr1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Judkins T, Hendrickson BC, Deffenbaugh AM, Eliason K, Leclair B, Norton MJ, Ward BE, Pruss D, Scholl T. Application of embryonic lethal or other obvious phenotypes to characterize the clinical significance of genetic variants found in trans with known deleterious mutations. Cancer Res. 2005;65:10096–10103. doi: 10.1158/0008-5472.CAN-05-1241. [DOI] [PubMed] [Google Scholar]
- 15.Couch FJ, Rasmussen LJ, Hofstra R, Monteiro AN, Greenblatt MS, de Wind N. Assessment of functional effects of unclassified genetic variants. Hum. Mutat. 2008;29:1314–1326. doi: 10.1002/humu.20899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fleming MA, Potter JD, Ramirez CJ, Ostrander GK, Ostrander EA. Understanding missense mutations in the BRCA1 gene: an evolutionary approach. Proc. Natl Acad. Sci. USA. 2003;100:1151–1156. doi: 10.1073/pnas.0237285100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Frederic MY, Lalande M, Boileau C, Hamroun D, Claustres M, Beroud C, Collod-Beroud G. UMD-predictor, a new prediction tool for nucleotide substitution pathogenicity—application to four genes: FBN1, FBN2, TGFBR1, and TGFBR2. Hum. Mutat. 2009;30:952–959. doi: 10.1002/humu.20970. [DOI] [PubMed] [Google Scholar]
- 18.Szabo C, Masiello A, Ryan JF, Brody LC. The breast cancer information core: database design, structure, and scope. Hum. Mutat. 2000;16:123–131. doi: 10.1002/1098-1004(200008)16:2<123::AID-HUMU4>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 19.Fokkema IF, den Dunnen JT, Taschner PE. LOVD: easy creation of a locus-specific sequence variation database using an “LSDB-in-a-box” approach. Hum. Mutat. 2005;26:63–68. doi: 10.1002/humu.20201. [DOI] [PubMed] [Google Scholar]
- 20.Osborne RH, Hopper JL, Kirk JA, Chenevix-Trench G, Thorne HJ, Sambrook JF. kConFab: a research resource of Australasian breast cancer families. Kathleen Cuningham Foundation Consortium for Research into Familial Breast Cancer. Med. J. Aust. 2000;172:463–464. doi: 10.5694/j.1326-5377.2000.tb124064.x. [DOI] [PubMed] [Google Scholar]
- 21.Tan EC, Loh M, Chuon D, Lim YP. Singapore human mutation/polymorphism database: a country-specific database for mutations and polymorphisms in inherited disorders and candidate gene association studies. Hum. Mutat. 2006;27:232–235. doi: 10.1002/humu.20291. [DOI] [PubMed] [Google Scholar]
- 22.Cooper DN, Stenson PD, Chuzhanova NA. The human gene mutation database (HGMD) and its exploitation in the study of mutational mechanisms. Curr. Protoc. Bioinformatics. 2006;Chapter 1:Unit 1 13. doi: 10.1002/0471250953.bi0113s12. [DOI] [PubMed] [Google Scholar]
- 23.Beroud C, Hamroun D, Collod-Beroud G, Boileau C, Soussi T, Claustres M. UMD (Universal Mutation Database): 2005 update. Hum. Mutat. 2005;26:184–191. doi: 10.1002/humu.20210. [DOI] [PubMed] [Google Scholar]
- 24.Beroud C, Collod-Beroud G, Boileau C, Soussi T, Junien C. UMD (Universal mutation database): a generic software to build and analyze locus-specific databases. Hum. Mutat. 2000;15:86–94. doi: 10.1002/(SICI)1098-1004(200001)15:1<86::AID-HUMU16>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 25.Cotton RG, Auerbach AD, Beckmann JS, Blumenfeld OO, Brookes AJ, Brown AF, Carrera P, Cox DW, Gottlieb B, Greenblatt MS, et al. Recommendations for locus-specific databases and their curation. Hum. Mutat. 2008;29:2–5. doi: 10.1002/humu.20650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Eisinger F, Alby N, Bremond A, Dauplat J, Espie M, Janiaud P, Kuttenn F, Lebrun JP, Lefranc JP, Pierret J, et al. Inserm ad hoc committee: recommendations for the management of women with a genetic risk for developing cancer of the breast and/or the ovary. Bull. Cancer. 1999;86:307–313. [PubMed] [Google Scholar]
- 27.Plon SE, Eccles DM, Easton D, Foulkes WD, Genuardi M, Greenblatt MS, Hogervorst FB, Hoogerbrugge N, Spurdle AB, Tavtigian SV. Sequence variant classification and reporting: recommendations for improving the interpretation of cancer susceptibility genetic test results. Hum. Mutat. 2008;29:1282–1291. doi: 10.1002/humu.20880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schouten JP, McElgunn CJ, Waaijer R, Zwijnenburg D, Diepvens F, Pals G. Relative quantification of 40 nucleic acid sequences by multiplex ligation-dependent probe amplification. Nucleic Acids Res. 2002;30:e57. doi: 10.1093/nar/gnf056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Casilli F, Di Rocco ZC, Gad S, Tournier I, Stoppa-Lyonnet D, Frebourg T, Tosi M. Rapid detection of novel BRCA1 rearrangements in high-risk breast-ovarian cancer families using multiplex PCR of short fluorescent fragments. Hum. Mutat. 2002;20:218–226. doi: 10.1002/humu.10108. [DOI] [PubMed] [Google Scholar]
- 30.Barrois M, Bieche I, Mazoyer S, Champeme MH, Bressac-de Paillerets B, Lidereau R. Real-time PCR-based gene dosage assay for detecting BRCA1 rearrangements in breast-ovarian cancer families. Clin. Genet. 2004;65:131–136. doi: 10.1111/j.0009-9163.2004.00200.x. [DOI] [PubMed] [Google Scholar]
- 31.Rouleau E, Lefol C, Bourdon V, Coulet F, Noguchi T, Soubrier F, Bieche I, Olschwang S, Sobol H, Lidereau R. Quantitative PCR high-resolution melting (qPCR-HRM) curve analysis, a new approach to simultaneously screen point mutations and large rearrangements: application to MLH1 germline mutations in Lynch syndrome. Hum. Mutat. 2009;30:867–875. doi: 10.1002/humu.20947. [DOI] [PubMed] [Google Scholar]
- 32.Weber J, Miserere S, Champ J, Looten R, Stoppa-Lyonnet D, Viovy JL, Houdayer C. High-throughput simultaneous detection of point mutations and large-scale rearrangements by CE. Electrophoresis. 2007;28:4282–4288. doi: 10.1002/elps.200700010. [DOI] [PubMed] [Google Scholar]
- 33.Gad S, Aurias A, Puget N, Mairal A, Schurra C, Montagna M, Pages S, Caux V, Mazoyer S, Bensimon A, et al. Color bar coding the BRCA1 gene on combed DNA: a useful strategy for detecting large gene rearrangements. Genes Chromosomes Cancer. 2001;31:75–84. doi: 10.1002/gcc.1120. [DOI] [PubMed] [Google Scholar]
- 34.Rouleau E, Lefol C, Tozlu S, Andrieu C, Guy C, Copigny F, Nogues C, Bieche I, Lidereau R. High-resolution oligonucleotide array-CGH applied to the detection and characterization of large rearrangements in the hereditary breast cancer gene BRCA1. Clin. Genet. 2007;72:199–207. doi: 10.1111/j.1399-0004.2007.00849.x. [DOI] [PubMed] [Google Scholar]
- 35.Tavtigian SV, Deffenbaugh AM, Yin L, Judkins T, Scholl T, Samollow PB, de Silva D, Zharkikh A, Thomas A. Comprehensive statistical study of 452 BRCA1 missense substitutions with classification of eight recurrent substitutions as neutral. J. Med. Genet. 2006;43:295–305. doi: 10.1136/jmg.2005.033878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ng PC, Henikoff S. SIFT: predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003;31:3812–3814. doi: 10.1093/nar/gkg509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ramensky V, Bork P, Sunyaev S. Human non-synonymous SNPs: server and survey. Nucleic Acids Res. 2002;30:3894–3900. doi: 10.1093/nar/gkf493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stoppa-Lyonnet D, Laurent-Puig P, Essioux L, Pages S, Ithier G, Ligot L, Fourquet A, Salmon RJ, Clough KB, Pouillart P, et al. BRCA1 sequence variations in 160 individuals referred to a breast/ovarian family cancer clinic. Institut Curie Breast Cancer Group. Am. J. Hum. Genet. 1997;60:1021–1030. [PMC free article] [PubMed] [Google Scholar]
- 39.Muller D, Bonaiti-Pellie C, Abecassis J, Stoppa-Lyonnet D, Fricker JP. BRCA1 testing in breast and/or ovarian cancer families from northeastern France identifies two common mutations with a founder effect. Fam. Cancer. 2004;3:15–20. doi: 10.1023/B:FAME.0000026819.44213.df. [DOI] [PubMed] [Google Scholar]
- 40.Hall MJ, Reid JE, Burbidge LA, Pruss D, Deffenbaugh AM, Frye C, Wenstrup RJ, Ward BE, Scholl TA, Noll WW. BRCA1 and BRCA2 mutations in women of different ethnicities undergoing testing for hereditary breast-ovarian cancer. Cancer. 2009;115:2222–2233. doi: 10.1002/cncr.24200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Suzuki K, Iwata M, Tsuji H, Takagi T, Tamura A, Ishimoto G, Ito S, Matsui K, Miyazaki T. A de novo recombination in the ABO blood group gene and evidence for the occurence of recombination products. Hum. Genet. 1997;99:454–461. doi: 10.1007/s004390050388. [DOI] [PubMed] [Google Scholar]
- 42.Diez O, Osorio A, Duran M, Martinez-Ferrandis JI, de la Hoya M, Salazar R, Vega A, Campos B, Rodriguez-Lopez R, Velasco E, et al. Analysis of BRCA1 and BRCA2 genes in Spanish breast/ovarian cancer patients: a high proportion of mutations unique to Spain and evidence of founder effects. Hum. Mutat. 2003;22:301–312. doi: 10.1002/humu.10260. [DOI] [PubMed] [Google Scholar]
- 43.Machado PM, Brandao RD, Cavaco BM, Eugenio J, Bento S, Nave M, Rodrigues P, Fernandes A, Vaz F. Screening for a BRCA2 rearrangement in high-risk breast/ovarian cancer families: evidence for a founder effect and analysis of the associated phenotypes. J. Clin. Oncol. 2007;25:2027–2034. doi: 10.1200/JCO.2006.06.9443. [DOI] [PubMed] [Google Scholar]
- 44.Petrij-Bosch A, Peelen T, van Vliet M, van Eijk R, Olmer R, Drusedau M, Hogervorst FB, Hageman S, Arts PJ, Ligtenberg MJ, et al. BRCA1 genomic deletions are major founder mutations in Dutch breast cancer patients. Nat. Genet. 1997;17:341–345. doi: 10.1038/ng1197-341. [DOI] [PubMed] [Google Scholar]
- 45.Palomba G, Pisano M, Cossu A, Budroni M, Dedola MF, Farris A, Contu A, Baldinu P, Tanda F, Palmieri G. Spectrum and prevalence of BRCA1 and BRCA2 germline mutations in Sardinian patients with breast carcinoma through hospital-based screening. Cancer. 2005;104:1172–1179. doi: 10.1002/cncr.21298. [DOI] [PubMed] [Google Scholar]
- 46.Hohenstein P, Kielman MF, Breukel C, Bennett LM, Wiseman R, Krimpenfort P, Cornelisse C, van Ommen GJ, Devilee P, Fodde R. A targeted mouse Brca1 mutation removing the last BRCT repeat results in apoptosis and embryonic lethality at the headfold stage. Oncogene. 2001;20:2544–2550. doi: 10.1038/sj.onc.1204363. [DOI] [PubMed] [Google Scholar]
- 47.Gowen LC, Johnson BL, Latour AM, Sulik KK, Koller BH. Brca1 deficiency results in early embryonic lethality characterized by neuroepithelial abnormalities. Nat. Genet. 1996;12:191–194. doi: 10.1038/ng0296-191. [DOI] [PubMed] [Google Scholar]
- 48.Evers B, Jonkers J. Mouse models of BRCA1 and BRCA2 deficiency: past lessons, current understanding and future prospects. Oncogene. 2006;25:5885–5897. doi: 10.1038/sj.onc.1209871. [DOI] [PubMed] [Google Scholar]
- 49.Howlett NG, Taniguchi T, Olson S, Cox B, Waisfisz Q, De Die-Smulders C, Persky N, Grompe M, Joenje H, Pals G, et al. Biallelic inactivation of BRCA2 in Fanconi anemia. Science. 2002;297:606–609. doi: 10.1126/science.1073834. [DOI] [PubMed] [Google Scholar]
- 50.Alter BP, Rosenberg PS, Brody LC. Clinical and molecular features associated with biallelic mutations in FANCD1/BRCA2. J. Med. Genet. 2007;44:1–9. doi: 10.1136/jmg.2006.043257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ludwig T, Chapman DL, Papaioannou VE, Efstratiadis A. Targeted mutations of breast cancer susceptibility gene homologs in mice: lethal phenotypes of Brca1, Brca2, Brca1/Brca2, Brca1/p53, and Brca2/p53 nullizygous embryos. Genes Dev. 1997;11:1226–1241. doi: 10.1101/gad.11.10.1226. [DOI] [PubMed] [Google Scholar]
- 52.Leegte B, van der Hout AH, Deffenbaugh AM, Bakker MK, Mulder IM, ten Berge A, Leenders EP, Wesseling J, de Hullu J, Hoogerbrugge N, et al. Phenotypic expression of double heterozygosity for BRCA1 and BRCA2 germline mutations. J. Med. Genet. 2005;42:e20. doi: 10.1136/jmg.2004.027243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Simard J, Dumont M, Moisan AM, Gaborieau V, Malouin H, Durocher F, Chiquette J, Plante M, Avard D, Bessette P, et al. Evaluation of BRCA1 and BRCA2 mutation prevalence, risk prediction models and a multistep testing approach in French-Canadian families with high risk of breast and ovarian cancer. J. Med. Genet. 2007;44:107–121. doi: 10.1136/jmg.2006.044388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Williams RS, Green R, Glover JN. Crystal structure of the BRCT repeat region from the breast cancer-associated protein BRCA1. Nat. Struct. Biol. 2001;8:838–842. doi: 10.1038/nsb1001-838. [DOI] [PubMed] [Google Scholar]
- 55.Shiozaki EN, Gu L, Yan N, Shi Y. Structure of the BRCT repeats of BRCA1 bound to a BACH1 phosphopeptide: implications for signaling. Mol. Cell. 2004;14:405–412. doi: 10.1016/s1097-2765(04)00238-2. [DOI] [PubMed] [Google Scholar]
- 56.Yang H, Jeffrey PD, Miller J, Kinnucan E, Sun Y, Thoma NH, Zheng N, Chen PL, Lee WH, Pavletich NP. BRCA2 function in DNA binding and recombination from a BRCA2-DSS1-ssDNA structure. Science. 2002;297:1837–1848. doi: 10.1126/science.297.5588.1837. [DOI] [PubMed] [Google Scholar]
- 57.Karchin R, Monteiro AN, Tavtigian SV, Carvalho MA, Sali A. Functional impact of missense variants in BRCA1 predicted by supervised learning. PLoS Comput. Biol. 2007;3:e26. doi: 10.1371/journal.pcbi.0030026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vallon-Christersson J, Cayanan C, Haraldsson K, Loman N, Bergthorsson JT, Brondum-Nielsen K, Gerdes AM, Moller P, Kristoffersson U, Olsson H, et al. Functional analysis of BRCA1 C-terminal missense mutations identified in breast and ovarian cancer families. Hum. Mol. Genet. 2001;10:353–360. doi: 10.1093/hmg/10.4.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Williams RS, Chasman DI, Hau DD, Hui B, Lau AY, Glover JN. Detection of protein folding defects caused by BRCA1-BRCT truncation and missense mutations. J. Biol. Chem. 2003;278:53007–53016. doi: 10.1074/jbc.M310182200. [DOI] [PubMed] [Google Scholar]
- 60.Nikolopoulos G, Pyrpassopoulos S, Thanassoulas A, Klimentzou P, Zikos C, Vlassi M, Vorgias CE, Yannoukakos D, Nounesis G. Thermal unfolding of human BRCA1 BRCT-domain variants. Biochim. Biophys. Acta. 2007;1774:772–780. doi: 10.1016/j.bbapap.2007.03.018. [DOI] [PubMed] [Google Scholar]
- 61.Karchin R, Agarwal M, Sali A, Couch F, Beattie MS. Classifying variants of undetermined significance in BRCA2 with protein likelihood ratios. Cancer Inform. 2008;6:203–216. doi: 10.4137/cin.s618. [DOI] [PMC free article] [PubMed] [Google Scholar]