Abstract
Biology is generating more data than ever. As a result, there is an ever increasing number of publicly available databases that analyse, integrate and summarize the available data, providing an invaluable resource for the biological community. As this trend continues, there is a pressing need to organize, catalogue and rate these resources, so that the information they contain can be most effectively exploited. MetaBase (MB) (http://MetaDatabase.Org) is a community-curated database containing more than 2000 commonly used biological databases. Each entry is structured using templates and can carry various user comments and annotations. Entries can be searched, listed, browsed or queried. The database was created using the same MediaWiki technology that powers Wikipedia, allowing users to contribute on many different levels. The initial release of MB was derived from the content of the 2007 Nucleic Acids Research (NAR) Database Issue. Since then, approximately 100 databases have been manually collected from the literature, and users have added information for over 240 databases. MB is synchronized annually with the static Molecular Biology Database Collection provided by NAR. To date, there have been 19 significant contributors to the project; each one is listed as an author here to highlight the community aspect of the project.
INTRODUCTION
When discussing biological databases, there are simply too many different resources to comprehensively cover the topic in a short introduction. There are well-established data warehouses that act as community repositories for data of a single type such as GenBank (1), PDB (2) and ArrayExpress (3). There are organism-specific databases, combining many different types of data under a unifying, genomic framework such as TAIR (4), FlyBase (5) and WormBase (6). There are databases of derived data, collecting and systematizing the body of knowledge from the scientific literature such as GTEx (http://www.ncbi.nlm.nih.gov/gtex/GTEX2/gtex.cgi), TRANSFAC (7), Brenda (8) and ChEMBL (9). There are competing databases that cover specific kinds of -omics information, collecting data from different experiments within a common biological theme such as DIP (10), HPID (11) and IntAct (12). There are classification databases (13,14), databases of terminology (15,16), databases of protein families (17,18) and databases built around diseases (19) or taxonomic groups (20). This list barely scratches the surface, but gives a flavour of the number, types and diversity of biological databases.
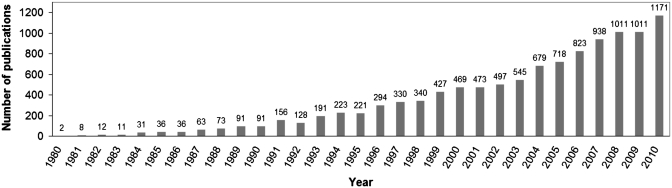
As the type and volume of biological data continues to increase, so do the type and number of databases that analyse, integrate and summarize the available data. For example, querying the database of biomedical publications PubMed (21) shows that the number of unique publications with the word ‘database’ in the title has increased from just 2 in 1980 to 91 in 1990 and 469 in 2000. Since 1990, there has been an exponential increase in the number of database publications per year, reaching over 1000 per year between 2008 and 2010 (Figure 1). If this trend continues, the number of database publications per year will double to nearly 2000 by 2015.
Figure 1.
The growth in the number of database publications per year. Each bar shows the number of research articles with the keyword ‘database’ appearing in the article title in the given year. The count only covers articles indexed in PubMed. The increase shows an exponential trend that will produce nearly 2000 database publications per year by 2015.
Biological databases have proven crucially important for basic research, however, the current growth in the available databases creates several problems. Researchers seeking the most up-to-date and comprehensive information in their domain may struggle to identify the definitive sources of reliable data from among the many resources available. Initially, it is difficult to judge the strengths, weaknesses, or status of the available resources without peer guidance. For these reasons, the proliferation of resources may, ironically, lead to an increase in redundancy, as new resources are created to cope with the perceived problems or omissions of existing databases. This process is exacerbated by a lack of public forums where researchers can engage database creators to discuss databases and suggest improvements.
These issues have created an unfortunate situation whereby many resources are short-lived, existing for only a short time before being abandoned. This ‘half-life’ is analogous to ‘link rot’ (22). This creates a vicious cycle, whereby the publication of database resources is devalued (23). To address these problems, we have created MetaBase (MB), a wiki-based database of biological databases.
DATABASE DESCRIPTION
MB is a community-curated database of all the biological databases available on the Internet. The aim of the project is to make it easy for researchers to quickly find relevant information about useful databases. Entries can be searched, queried or browsed by category, and users can contribute, update and maintain the data in many different ways. Each database in MB is described in a semi-structured way using forms and templates. Entries carry data for various fields and allow a free-text description of the resource. In detail, data for each database include a brief description, a URL, a contact email, links to associated literature and various categorization tags. In addition, entries can carry various user comments and annotations.
MB has been implemented using MediaWiki (MW), the same software that powers Wikipedia, probably the best known user-contributed resource in the world (http://wikipedia.org). The MediaWiki system allows users to contribute to the project on many different levels, ranging from authors and editors to curators and site designers. Within the MW system, we created one wiki-page per database entry. The information about each database is structured by using a template with named fields. The template stores data for each database internally using the Semantic MediaWiki extension (http://semantic-mediawiki.org), allowing data to be queried within the wiki directly, by additional extensions or via the semantic web. In particular, we use the Semantic Forms extension (http://www.mediawiki.org/wiki/SF) to allow users to create or edit entries and the Semantic Drilldown extension (http://www.mediawiki.org/wiki/SD) to allow users to explore the database. User comments are collected as free text, just like in Wikipedia.
FEATURES
The MW platform provides a robust base from which to build an online resource. By using MW, many powerful features are provided ‘for free’. The use of MW to support Wikipedia demonstrates the scalability and security of the system, guaranteeing developer support and providing a degree of familiarity to users. Out of the box, MW provides searching, editing, versioning, history and discussion features, as well as user account management and user-email functions. MW includes a powerful extension framework for easily adding functionality.
One criticism of MW is that it provides largely unstructured information, not suitable for advanced searching or reporting. To this end, we employ Semantic MediaWiki and Semantic Forms to create a wiki-database system suitable for maintaining a user-contributed database of information.
DATABASE CONTENTS
Currently, there are 1795 entries in MB, each describing a different biological database. The initial release was derived from the content of the 2007 Nucleic Acids Research (NAR) Database Issue (24). Specifically, each database page was ‘seeded’ with text from the Molecular Biology Database Collection provided by NAR (25). Subsequent releases have been updated into MB on a semi-regular basis. Since the initial release, there have been over 100 user contributed resources added, in addition to 100 resources that were manually collected from the literature. Most of these were taken from database publications in BMC Bioinformatics and BMC Biology. To date, there have been 19 significant contributors to the project, each of whom has been listed as an author on this publication. This step was taken to highlight the community aspect of the MB project. The homepage has been visited approximately 100 000 times. The project has 80 registered users in total, and there have been approximately 15 000 edits. We hope that with ongoing improvements and through increased publicity, usage will continue to grow helping to establish MB as a powerful and referential community resource.
FUTURE DIRECTIONS
In the future, we hope to use MB as a resource to allow more communication between database developers and user communities, acting as a common portal for the biological database community. To achieve this goal, we will automatically register the database's contact email address and add the database's discussion page to that user's ‘watch list’. Comments will then automatically alert the contact, providing them with the opportunity to reply. We hope to add user rating functionality and usage statistics to each resource. This will be done with a combination of existing MediaWiki extensions, adding links to social networking sites and automatic queries to collect the number of citations for each resource. We expect that MB could be used as a source of genuine metadata for data integration projects, and we plan to incorporate ontologies such as EDaM (26,27) and the Biomedical Resource Ontology (28), and to develop links with similar projects such as BioCatalogue (29) and BioDBCore (30).
Finally, we aim to improve the content of MB through an aggressive marketing strategy, contacting the relevant mailing lists, forums and news groups, as well as exploiting the collection of contact email addresses, thereby encouraging the community to contribute to the maintenance of this important resource.
RELATED WORK
MB is by no means unique. There are many related resources, falling into two broad categories: ‘BioWikis’ and ‘databases of biological databases’.
First, there are several other ‘BioWiki’ projects. Like MB, these projects use the tremendously successful MediaWiki software platform to provide user-contributed content to the biological community. For a comprehensive list of important and interesting BioWiki projects, see the BioWiki database on Bioinformatcs.Org (http://bioinformatics.org/wiki/BioWiki). The most successful collection of user-contributed content is Wikipedia (http://www.wikipedia.org/). The success of Wikipedia is intimately related to the success of the MediaWiki software platform, leading to a proliferation of wikis, including several BioWiki projects. However, Wikipedia is still a very important resource for biologists (e.g. http://en.wikipedia.org/wiki/Wikipedia:MCB). Wikipedia maintains a sizeable list of biological databases (http://en.wikipedia.org/wiki/List_of_biological_databases), and many of the databases in MB also have articles in Wikipedia.
Second, there are several ‘databases of biological databases’, which aim to provide a list of all the most important biological databases and data resources available on the Internet. Several prominent biological database collections and related projects are listed in Table 1 (see also http://metadatabase.org/wiki/Help:Related).
Table 1.
Projects with a similar scope to MB
| Name | Description | URL |
|---|---|---|
| The Molecular Biology Database Collection | A public on-line resource that lists the databases described in Nucleic Acids Research, together with other databases of value to the biologist (25). | http://www.oxfordjournals.org/nar/database/c/ |
| OBRC: Online Bioinformatics Resources Collection | Contains annotations and links for 1746 bioinformatics databases and software tools. | http://www.hsls.pitt.edu/guides/genetics/obrc/ |
| The Bioinformatics Links Directory | Features curated links to molecular resources, tools and databases (31). | http://bioinformatics.ca/links_directory/ |
| CABRI: Common Access to Biotechnology Resources and Information | An service to search European Biological Resource Centre catalogues. The catalogues may be searched independently, or as one, and the located materials ordered online or by post (32). | http://www.cabri.org/ |
| DBD: Database of Biological Database | Consists of 1200 database entries covering wide range of databases useful for biological researchers. | http://www.biodbs.info/ |
| BioDBCore | A community-defined description of the core attributes of biological databases (28). | http://biocurator.org/biodbcore.shtml |
| MetaBasis | A database of metadata for bioinformatics software tools and databases. The system contains 3229 published bioinformatics tools and databases (33). | http://bioserver-1.bioacademy.gr/Metabasis/ |
| Biomed Central Databases | A catalogue of online databases with more than 1100 sites covering a wide range of biomedical topics. | http://databases.biomedcentral.com/ |
| OReFiL | An Online Resource Finder for Life sciences (34). | http://orefil.dbcls.jp/ |
| NIST Data Gateway | Provides easy access to many of the The National Institute of Standards and Technology databases, covering a many different scientific disciplines. | http://srdata.nist.gov/gateway/ |
These projects aim to list the most important biological databases and data resources available on the Internet. For a version of this table that you can edit, see http://metadatabase.org/wiki/Help:Related
DISCUSSION
Biological databases have proven crucially important for basic research. However, exponential growth in the volume of biological data has led to several problems. MB is an international, community-based database that aims to list all the commonly used biological databases in the world. Here, we have created a new scientific-wiki that addresses some of the issues described earlier. The first version of the system was based on a static database of biological databases that has been imported to a wiki system for community annotation. Although similar to several other ‘lists of resources’, MB is unique, being the only truly user-editable list of databases. The NAR Molecular Biology Database Collection is a curated database with strict criteria for inclusion. It covers only a relatively small number of the available molecular biology databases (M. Galperin, personal communication). In contrast, we hope MB, with its liberal wiki-based inclusion policy, might be useful as a wider, more general list with quicker updates.
FUNDING
Industrial Strategic technology development program, (10040231), “Bioinformatics platform development for next generation bioinformation analysis” funded by the Ministry of Knowledge Economy (MKE, Korea). Funding for Open access charge: Genome Research Foundation's internal Biowiki funds.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
MB was first hosted on the Bioinformation Objects Community Cluster (BiO.CC) and was created as a competition entry hosted by BiO.CC. Currently, MB is hosted at Bioinformatics.Org. D.M.B. would like to thank J.B. and Jeff Bizzaro for hosting and all the contributors to MB.
REFERENCES
- 1.Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW. GenBank. Nucleic Acids Res. 2011;39:D32–D37. doi: 10.1093/nar/gkq1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rose PW, Beran B, Bi C, Bluhm WF, Dimitropoulos D, Goodsell DS, Prlic A, Quesada M, Quinn GB, Westbrook JD, et al. The RCSB Protein Data Bank: redesigned web site and web services. Nucleic Acids Res. 2011;39:D392–D401. doi: 10.1093/nar/gkq1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Parkinson H, Sarkans U, Kolesnikov N, Abeygunawardena N, Burdett T, Dylag M, Emam I, Farne A, Hastings E, Holloway E, et al. ArrayExpress update–an archive of microarray and high-throughput sequencing-based functional genomics experiments. Nucleic Acids Res. 2011;39:D1002–D1004. doi: 10.1093/nar/gkq1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Swarbreck D, Wilks C, Lamesch P, Berardini TZ, Garcia-Hernandez M, Foerster H, Li D, Meyer T, Muller R, Ploetz L, et al. The Arabidopsis Information Resource (TAIR): gene structure and function annotation. Nucleic Acids Res. 2008;36:D1009–D1014. doi: 10.1093/nar/gkm965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tweedie S, Ashburner M, Falls K, Leyland P, McQuilton P, Marygold S, Millburn G, Osumi-Sutherland D, Schroeder A, Seal R, et al. FlyBase Consortium FlyBase: enhancing Drosophila Gene Ontology annotations. Nucleic Acids Res. 2009;37:D555–D559. doi: 10.1093/nar/gkn788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rogers A, Antoshechkin I, Bieri T, Blasiar D, Bastiani C, Canaran P, Chan J, Chen WJ, Davis P, Fernandes J, et al. WormBase 2007. Nucleic Acids Res. 2008;36:D612–D617. doi: 10.1093/nar/gkm975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Knüppel R, Dietze P, Lehnberg W, Frech K, Wingender E. TRANSFAC retrieval program: a network model database of eukaryotic transcription regulating sequences and proteins. J. Comput. Biol. 1994;1:191–198. doi: 10.1089/cmb.1994.1.191. [DOI] [PubMed] [Google Scholar]
- 8.Scheer M, Grote A, Chang A, Schomburg I, Munaretto C, Rother M, Söhngen C, Stelzer M, Thiele J, Schomburg D. BRENDA, the enzyme information system in 2011. Nucleic Acids Res. 2011;39:D670–D676. doi: 10.1093/nar/gkq1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gaulton A, Bellis LJ, Bento AP, Chambers J, Davies M, Hersey A, Light Y, McGlinchey S, Michalovich D, Al-Lazikani B, et al. ChEMBL: a large-scale bioactivity database for drug discovery. Nucleic Acids Res. 2012;40:D1100–D1107. doi: 10.1093/nar/gkr777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Salwinski L, Miller CS, Smith AJ, Pettit FK, Bowie JU, Eisenberg D. The Database of Interacting Proteins: 2004 update. Nucleic Acids Res. 2004;32:D449–D451. doi: 10.1093/nar/gkh086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Han K, Park B, Kim H, Hong J, Park J. HPID: the Human Protein Interaction Database. Bioinformatics. 2004;20:2466–2470. doi: 10.1093/bioinformatics/bth253. [DOI] [PubMed] [Google Scholar]
- 12.Aranda B, Achuthan P, Alam-Faruque Y, Armean I, Bridge A, Derow C, Feuermann M, Ghanbarian AT, Kerrien S, Khadake J, et al. The IntAct molecular interaction database in 2010. Nucleic Acids Res. 2010;38:D525–D531. doi: 10.1093/nar/gkp878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Andreeva A, Howorth D, Chandonia JM, Brenner SE, Hubbard TJ, Chothia C, Murzin AG. Data growth and its impact on the SCOP database: new developments. Nucleic Acids Res. 2008;36:D419–D425. doi: 10.1093/nar/gkm993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cuff AL, Sillitoe I, Lewis T, Clegg AB, Rentzsch R, Furnham N, Pellegrini-Calace M, Jones D, Thornton J, Orengo CA. Extending CATH: increasing coverage of the protein structure universe and linking structure with function. Nucleic Acids Res. 2011;39:D420–D426. doi: 10.1093/nar/gkq1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bairoch A. The ENZYME database in 2000. Nucleic Acids Res. 2000;28:304–305. doi: 10.1093/nar/28.1.304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gene Ontology Consortium. The Gene Ontology in 2010: extensions and refinements. Nucleic Acids Res. 2010;38:D331–D335. doi: 10.1093/nar/gkp1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Finn RD, Mistry J, Tate J, Coggill P, Heger A, Pollington JE, Gavin OL, Gunasekaran P, Ceric G, Forslund K, et al. The Pfam protein families database. Nucleic Acids Res. 2010;38:D211–D222. doi: 10.1093/nar/gkp985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tatusov RL, Fedorova ND, Jackson JD, Jacobs AR, Kiryutin B, Koonin EV, Krylov DM, Mazumder R, Mekhedov SL, Nikolskaya AN, et al. The COG database: an updated version includes eukaryotes. BMC Bioinformatics. 2003;4:41. doi: 10.1186/1471-2105-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Forbes SA, Bindal N, Bamford S, Cole C, Kok CY, Beare D, Jia M, Shepherd R, Leung K, Menzies A, et al. COSMIC: mining complete cancer genomes in the catalogue of somatic mutations in cancer. Nucleic Acids Res. 2011;39: D945–D950. doi: 10.1093/nar/gkq929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bombarely A, Menda N, Tecle IY, Buels RM, Strickler S, Fischer-York T, Pujar A, Leto J, Gosselin J, Mueller LA. The Sol Genomics Network (solgenomics.net): growing tomatoes using Perl. Nucleic Acids Res. 2011;39:D1149–D1155. doi: 10.1093/nar/gkq866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McEntyre J, Lipman D. PubMed: bridging the information gap. CMAJ. 2001;164:1317–1319. [PMC free article] [PubMed] [Google Scholar]
- 22.Koehler W. A longitudinal study of Web pages continued: a report after six years. Informat. Res. 2004;9 paper 174. [Google Scholar]
- 23.Tan TW, Tong JC, Khan AK, de Silva M, Lim KS, Ranganathan S. Advancing standards for bioinformatics activities: persistence, reproducibility, disambiguation and Minimum Information About a Bioinformatics investigation (MIABi) BMC Genomics. 2010;11:S27. doi: 10.1186/1471-2164-11-S4-S27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bateman A. Editorial. Nucleic Acids Res. 2007;35:D1–D2. [Google Scholar]
- 25.Galperin MY, Cochrane GR. The 2011 Nucleic Acids Research Database Issue and the online Molecular Biology Database Collection. Nucleic Acids Res. 2011;39:D1–D6. doi: 10.1093/nar/gkq1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pettifer S, Ison J, Kalas M, Thorne D, McDermott P, Jonassen I, Liaquat A, Fernández JM, Rodriguez JM, et al. INB-Partners The EMBRACE web service collection. Nucleic Acids Res. 2010;38:W683–W688. doi: 10.1093/nar/gkq297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kalas M, Puntervoll P, Joseph A, Bartaseviciūte E, Töpfer A, Venkataraman P, Pettifer S, Bryne JC, Ison J, Blanchet C, et al. BioXSD: the common data-exchange format for everyday bioinformatics web services. Bioinformatics. 2010;26:i540–i546. doi: 10.1093/bioinformatics/btq391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tenenbaum JD, Whetzel PL, Anderson K, Borromeo CD, Dinov ID, Gabriel D, Kirschner B, Mirel B, Morris T, Noy N, et al. The Biomedical Resource Ontology (BRO) to enable resource discovery in clinical and translational research. J. Biomed. Inform. 2011;44:137–145. doi: 10.1016/j.jbi.2010.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bhagat J, Tanoh F, Nzuobontane E, Laurent T, Orlowski J, Roos M, Wolstencroft K, Aleksejevs S, Stevens R, Pettifer S, et al. BioCatalogue: a universal catalogue of web services for the life sciences. Nucleic Acids Res. 2010;38:W689–W694. doi: 10.1093/nar/gkq394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gaudet P, Bairoch A, Field D, Sansone S-A, Taylor C, Attwood TK, Bateman A, Blake JA, Bult CJ, Cherry JM, et al. Towards BioDBcore: a community-defined information specification for biological databases. Nucleic Acids Res. 2011;39:D7–D10. doi: 10.1093/nar/gkq1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen YB, Chattopadhyay A, Bergen P, Gadd C, Tannery N. The Online Bioinformatics Resources Collection at the University of Pittsburgh Health Sciences Library System–a one-stop gateway to online bioinformatics databases and software tools. Nucleic Acids Res. 2007;35:D780–D785. doi: 10.1093/nar/gkl781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Romano P, Aresu O, Manniello M, Parodi B. Interoperability of CABRI Services and Biochemical Pathways Databases. Comp. Funct. Genomics. 2004;5:169–172. doi: 10.1002/cfg.376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Atlamazoglou V, Thireou T, Hamodrakas Y, Spyrou G. MetaBasis: a web-based database containing metadata on software tools and databases in the field of bioinformatics. Appl. Bioinformatics. 2006;5:187–192. doi: 10.2165/00822942-200605030-00007. [DOI] [PubMed] [Google Scholar]
- 34.Yamamoto Y, Takagi T. OReFiL: an online resource finder for life sciences. BMC Bioinformatics. 2007;8:287. doi: 10.1186/1471-2105-8-287. [DOI] [PMC free article] [PubMed] [Google Scholar]