Abstract
RNA splicing is an important aspect of gene regulation in many organisms. Splicing of RNA is regulated by complicated mechanisms involving numerous RNA-binding proteins and the intricate network of interactions among them. Mutations in cis-acting splicing elements or its regulatory proteins have been shown to be involved in human diseases. Defects in pre-mRNA splicing process have emerged as a common disease-causing mechanism. Therefore, a database integrating RNA splicing and disease associations would be helpful for understanding not only the RNA splicing but also its contribution to disease. In SpliceDisease database, we manually curated 2337 splicing mutation disease entries involving 303 genes and 370 diseases, which have been supported experimentally in 898 publications. The SpliceDisease database provides information including the change of the nucleotide in the sequence, the location of the mutation on the gene, the reference Pubmed ID and detailed description for the relationship among gene mutations, splicing defects and diseases. We standardized the names of the diseases and genes and provided links for these genes to NCBI and UCSC genome browser for further annotation and genomic sequences. For the location of the mutation, we give direct links of the entry to the respective position/region in the genome browser. The users can freely browse, search and download the data in SpliceDisease at http://cmbi.bjmu.edu.cn/sdisease.
INTRODUCTION
Cells need to regulate the expression of a gene in a specific level at specific time and space in order to fulfill specific task. Gene regulation is a ubiquitous phenomenon and is critical in every biological process (1). Mechanisms of gene regulation include the regulations of transcription, RNA processing and translation. In higher eukaryotes, pre-mRNA splicing plays an important role in gene regulation. The inclusion of different exons in mRNA—alternative splicing (AS)—enables one single gene to produce multiple different mRNAs, which can be further translated into different proteins called splice variants (2,3). New high-throughput sequencing technology has revealed that >90% of human genes undergo AS—a much higher percentage than anticipated (4). And recent genome-wide analyses have indicated that almost all primary transcripts from multi-exon human genes undergo alternative pre-mRNA splicing (5). Therefore, RNA splicing greatly increases the genomic complexity of higher eukaryotes (6). RNA splicing is tissue specific and studies highlight differences in the types of AS occurring commonly in different tissues. For example, the frequencies of alternative 3′ splice site and alternative 5′ splice site usage are ∼50–100% higher in liver than in other investigated tissues (7). The importance of splicing is emphasized by its presence in species throughout the phylogenetic tree. Evolutionary studies, which have revealed the formation of de novo alternative exons and the evolution of exon–intron architecture, highlight the importance of AS in the diversification of the transcriptome, especially in humans (8).
As we stated earlier, RNA splicing is critical in many biological processes. Splicing of RNA is regulated by complicated mechanisms involving numerous RNA-binding proteins and the intricate network of interactions among them. Splicing in general, and AS in particular, if disrupted, can lead to disease. Therefore, mutations in cis-acting splicing elements or splicing machinery and the regulatory proteins which could compromise the accuracy of either constitutive or alternative splicing would have a profound impact on human pathogenesis. Defects in pre-mRNA splicing have been shown as a common disease-causing mechanism in several studies (9–11). As an example, a point mutation in exon 7 of SMN2 gene leads to exon 7 skipping and a truncated protein, which causes decreased effective rate of SMN protein production and motor neuron degenerative disease (12). Other studies indicate trans-acting mutations affect RNA-dependent functions and cause disease (9,13). A number of bioinformatics resources for RNA splicing have been developed during the past decade including databases and tools (Table 1). For example, Human Splicing Finder is a tool to predict the effects of mutations on splicing signals and can identify splicing motifs in human sequence (14). These resources have provided great help in the study and analysis of RNA splicing.
Table 1.
Databases and tools of splicing mutation and alternative splicing
| Resource | Description | URL |
|---|---|---|
| HGMD (15) | The Human Gene Mutation Database (HGMD) constitutes a comprehensive core collection of data on germ-line mutations in nuclear genes underlying or associated with human inherited disease | www.hgmd.org |
| DBASS5 (16,17) | A database of aberrant 5′ splice sites | http://www.dbass.org.uk/ |
| DBASS3 (17,18) | A database of aberrant 3′ splice sites | http://www.dbass.org.uk/ |
| ASDB (19) | Database of alternatively spliced genes | http://cbcg.nersc.gov/asdb |
| ssSNPTarget (20) | A genome-wide splice-site Single Nucleotide Polymorphism database | http://ssSNPTarget.org |
| EuSplice (21) | a splice-centric database which provides reliable splice signal and AS information for 23 eukaryotes | http://66.170.16.154/EuSplice |
| AsMamDB (22) | An alternative splice database of mammals | http://166.111.30.65/ASMAMDB.html |
| Alternative Splicing Database (23) | An alternative splicing database based on publications | http://cgsigma.cshl.org/new_alt_exon_db2/ |
| TassDB2 (24) | A database of subtle alternative splicing events | http://www.tassdb.info |
| ISIS (25) | An intron information system | http://isis.bit.uq.edu.au/ |
| ASPicDB (26) | A database of annotated transcript and protein variants generated by alternative splicing | http://www.caspur.it/ASPicDB/ |
| STEPs (27) | A database of splice translational efficiency polymorphisms | http://dbstep.genes.org.uk/ |
| Human Splicing Finder (14) | A tool to predict the effects of mutations on splicing signals or to identify splicing motifs in any human sequence | http://www.umd.be/HSF/ |
| SpliceMiner (28) | A high-throughput database implementation of the NCBI Evidence Viewer for microarray splice variant analysis | http://discover.nci.nih.gov/spliceminer |
| Intronerator (29) | Exploring introns and alternative splicing in Caenorhabditis elegans | http://www.cse.ucsc.edu/~kent/intronerator |
| WebScipio (30) | Tool for Predicting mutually exclusive spliced exons based on exon length, splice site and reading frame conservation, and exon sequence homology | http://www.webscipio.org |
| IsoEM (31) | Tool for the estimation of alternative splicing isoform frequencies from RNA-Seq data | http://dna.engr.uconn.edu/software/IsoEM/ |
| MAISTAS (32) | A tool for automatic structural evaluation of alternative splicing products | http://maistas.bioinformatica.crs4.it/ |
| HMMSplicer (33) | A tool for efficient and sensitive discovery of known and novel splice junctions in RNA-Seq data | http://derisilab.ucsf.edu/software/hmmsplicer |
| SFmap (34) | A web server for motif analysis and prediction of splicing factor binding sites | http://sfmap.technion.ac.il |
The above evidences have shown an increased importance of connecting the RNA splicing and diseases. For this reason, a high quality database linking RNA splicing and splicing mutations with disease will be of great help and be emergently needed in the study of both RNA splicing and disease. Although the human gene mutation database (HGMD, http://www.hgmd.org/) integrated this kind of data but there are big difference between HGMD and SpliceDisease database (15). Firstly, HGMD is not free and only provides ‘search’ function for registered users for limited days. Secondly, HGMD only provides information of point mutations of intronic sequence for splicing mutation. Thirdly, HGMD does not provide detailed descriptions for the relationship among gene mutations, splicing defects and diseases. On the other hand, SpliceDisease database is a free and comprehensive database containing cis-splicing sequence mutations and trans-acting splicing mutations that cause disease. SpliceDisease integrates detailed descriptions for the relationship among gene mutation, splicing defect and disease. And it provides direct links of EntreZ gene, genome browser, respective location of the mutation on the gene and PubMed for each literature. At present, the ‘SpliceDisease’ database is at its first step, it will be a valuable ongoing resource for the study of RNA splicing and disease.
DATA SOURCES AND IMPLEMENTATION
RNA splicing and disease-related literature was acquired by PubMed search using the keywords ‘splice’, ‘splicing’ and ‘spliced’. Literatures with titles including ‘mutation spectrum’, ‘mutational spectrum’, ‘mutation analysis’ and ‘mutation screening’ were also obtained. We then curated the data manually and retrieved the association between RNA splicing and splicing mutations in the gene and disease of interest. The data were double checked by different people. We standardized the disease names and gene names based on NLM Mesh Browser and EntreZ gene. Each gene was linked to NCBI for comprehensive annotations and to UCSC genome browser for genomic sequence. The mutations of genes were annotated as well including nucleotide change and location on the sequence. We used the nomenclature for description of sequence variants and exon/intron numbering according to den Dunnen and Antonarakis (35). For example: ‘c.’ for a cDNA sequence; IVS for intron sequence; substitutions are designated by a ‘>’ character. We also gave direct link for entry to the respective position of mutation. In the sequence file, the intron/exon of the mutation location is highlighted in yellow color and the specific nucleotide is marked in red color. PubMed IDs and hyperlinks to PubMed were also provided for each literature. More importantly, we curated the detailed description for the relationship among gene mutation, splicing defect and disease.
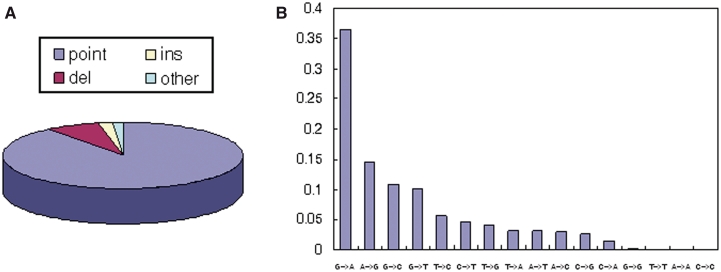
As a result, we manually curated 2337 splicing mutation-disease entries including 303 genes and 370 diseases from 898 publications. In the 2337 entries, ∼89% of them are point mutations (Figure 1A) among which >50% are mutations between G and A (36.5% G > A and 14.6% A > G) (Figure 1B).
Figure 1.
Distribution of mutation type and distribution of point mutation type in the SpliceDisease database. (A) Splicing mutation type: point, point mutation; ins, insertion mutation; del, deletion mutation; other, other types. (B) Axes of the histogram represent the proportions of different nucleotide substitutions in whole point mutations.
All data were organized in the ‘SpliceDisease’ database using PostgreSQL 9.0, a lightweight database management system. The website is presented using Apache Tomcat 7.0, a JSP&Java web framework which is available at http://cmbi.bjmu.edu.cn/sdisease/.
USING SpliceDisease
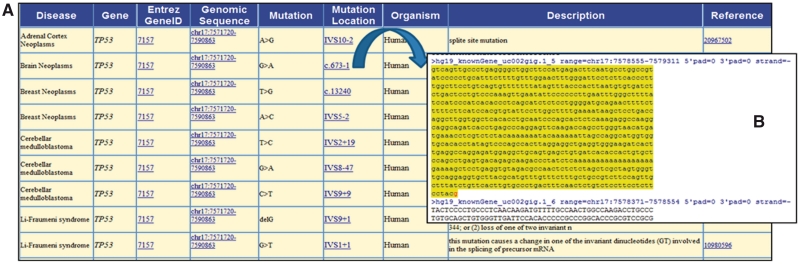
SpliceDisease is a user-friendly designed database. The homepage has been designed to provide an organized venue to access all data. The top banner section of the homepage has tabs for ‘Browser’, ‘Search’, ‘Submit’, ‘Download’ and ‘Help’, respectively. When a user performs a search in SpliceDisease, he can use the ‘Browser’ to select the disease or gene of interest or use the ‘Search’ which supports fuzzy queries to find it. The page of result contains nine items disease name and gene symbol, gene EntreZ ID (link to NCBI gene database), chromosome location of genomic sequence (link to UCSC genome browser), mutation, mutation location (direct link to respective position of mutation in the genome browser automatically), organism, description and reference (link to PubMed database) (Figure 2).
Figure 2.
SpliceDisease results page. (A) Once a user runs a search, there comes the result summary page that includes nine items. (B) The direct link for entry to the respective position of mutation. The sequence of exon shows in upper case and intron shows in lower case. And one FASTA record per region (exon, intron) is used in the sequence file. The inton/exon of the location of mutation is highlighted in yellow color and specific nucleotide is marked in red color.
All data in SpliceDisease can be downloaded in a file of csv format. These data will facilitate study of exploitation of splicing mutational mechanisms, understanding of RNA biology and helping to discover new therapeutic targets.
FUTURE EXTENSIONS
The SpliceDisease database is in the first step of the project and further extensions will be developed. As we described earlier, a number of bioinformatics resources for RNA splicing have been developed. Therefore, we plan to integrate some related bioinformatics resources in the near future. We will also incorporate the expression data of different mRNA isoforms. As the data accumulation, we will add more trans-acting splicing mutations that cause disease. Finally, SpliceDisease will be continuously updated.
FUNDING
Funding for open access charge: National Natural Science Foundation of China (Grant No. 81001481).
Conflict of interest statement. None declared.
REFERENCES
- 1.Holste D, Ohler U. Strategies for identifying RNA splicing regulatory motifs and predicting alternative splicing events. PLoS Comput. Biol. 2008;4:e21. doi: 10.1371/journal.pcbi.0040021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Xing Y, Resch A, Lee C. The multiassembly problem: reconstructing multiple transcript isoforms from EST fragment mixtures. Genome Res. 2004;14:426–441. doi: 10.1101/gr.1304504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Modrek B, Lee C. A genomic view of alternative splicing. Nat. Genet. 2002;30:13–19. doi: 10.1038/ng0102-13. [DOI] [PubMed] [Google Scholar]
- 4.Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Johnson JM, Castle J, Garrett-Engele P, Kan Z, Loerch PM, Armour CD, Santos R, Schadt EE, Stoughton R, Shoemaker DD. Genome-wide survey of human alternative pre-mRNA splicing with exon junction microarrays. Science. 2003;302:2141–2144. doi: 10.1126/science.1090100. [DOI] [PubMed] [Google Scholar]
- 6.Brett D, Pospisil H, Valcarcel J, Reich J, Bork P. Alternative splicing and genome complexity. Nat. Genet. 2002;30:29–30. doi: 10.1038/ng803. [DOI] [PubMed] [Google Scholar]
- 7.Yeo G, Holste D, Kreiman G, Burge CB. Variation in alternative splicing across human tissues. Genome Biol. 2004;5:R74. doi: 10.1186/gb-2004-5-10-r74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Keren H, Lev-Maor G, Ast G. Alternative splicing and evolution: diversification, exon definition and function. Nat. Rev. Genet. 2010;11:345–355. doi: 10.1038/nrg2776. [DOI] [PubMed] [Google Scholar]
- 9.Cooper TA, Wan L, Dreyfuss G. RNA and disease. Cell. 2009;136:777–793. doi: 10.1016/j.cell.2009.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Faustino NA, Cooper TA. Pre-mRNA splicing and human disease. Genes Dev. 2003;17:419–437. doi: 10.1101/gad.1048803. [DOI] [PubMed] [Google Scholar]
- 11.Tazi J, Bakkour N, Stamm S. Alternative splicing and disease. Biochim. Biophys. Acta. 2009;1792:14–26. doi: 10.1016/j.bbadis.2008.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wirth B, Brichta L, Hahnen E. Spinal muscular atrophy: from gene to therapy. Semin. Pediatr. Neurol. 2006;13:121–131. doi: 10.1016/j.spen.2006.06.008. [DOI] [PubMed] [Google Scholar]
- 13.Liu F, Gong CX. Tau exon 10 alternative splicing and tauopathies. Mol. Neurodegener. 2008;3:8. doi: 10.1186/1750-1326-3-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Desmet FO, Hamroun D, Lalande M, Collod-Beroud G, Claustres M, Beroud C. Human Splicing Finder: an online bioinformatics tool to predict splicing signals. Nucleic Acids Res. 2009;37:e67. doi: 10.1093/nar/gkp215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cooper DN, Stenson PD, Chuzhanova NA. The Human Gene Mutation Database (HGMD) and its exploitation in the study of mutational mechanisms. Curr. Protoc. Bioinform. 2006 doi: 10.1002/0471250953.bi0113s12. Chapter 1, Unit 1 13. [DOI] [PubMed] [Google Scholar]
- 16.Buratti E, Chivers M, Kralovicova J, Romano M, Baralle M, Krainer AR, Vorechovsky I. Aberrant 5′ splice sites in human disease genes: mutation pattern, nucleotide structure and comparison of computational tools that predict their utilization. Nucleic Acids Res. 2007;35:4250–4263. doi: 10.1093/nar/gkm402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Buratti E, Chivers M, Hwang G, Vorechovsky I. DBASS3 and DBASS5: databases of aberrant 3′- and 5′-splice sites. Nucleic Acids Res. 2011;39:D86–D91. doi: 10.1093/nar/gkq887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vorechovsky I. Aberrant 3′ splice sites in human disease genes: mutation pattern, nucleotide structure and comparison of computational tools that predict their utilization. Nucleic Acids Res. 2006;34:4630–4641. doi: 10.1093/nar/gkl535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dralyuk I, Brudno M, Gelfand MS, Zorn M, Dubchak I. ASDB: database of alternatively spliced genes. Nucleic Acids Res. 2000;28:296–297. doi: 10.1093/nar/28.1.296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang JO, Kim WY, Bhak J. ssSNPTarget: genome-wide splice-site Single Nucleotide Polymorphism database. Hum. Mutat. 2009;30:E1010–E1020. doi: 10.1002/humu.21128. [DOI] [PubMed] [Google Scholar]
- 21.Bhasi A, Pandey RV, Utharasamy SP, Senapathy P. EuSplice: a unified resource for the analysis of splice signals and alternative splicing in eukaryotic genes. Bioinformatics. 2007;23:1815–1823. doi: 10.1093/bioinformatics/btm084. [DOI] [PubMed] [Google Scholar]
- 22.Ji H, Zhou Q, Wen F, Xia H, Lu X, Li Y. AsMamDB: an alternative splice database of mammals. Nucleic Acids Res. 2001;29:260–263. doi: 10.1093/nar/29.1.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stamm S, Zhu J, Nakai K, Stoilov P, Stoss O, Zhang MQ. An alternative-exon database and its statistical analysis. DNA Cell Biol. 2000;19:739–756. doi: 10.1089/104454900750058107. [DOI] [PubMed] [Google Scholar]
- 24.Sinha R, Lenser T, Jahn N, Gausmann U, Friedel S, Szafranski K, Huse K, Rosenstiel P, Hampe J, Schuster S, et al. TassDB2 - A comprehensive database of subtle alternative splicing events. BMC Bioinformatics. 2010;11:216. doi: 10.1186/1471-2105-11-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Croft L, Schandorff S, Clark F, Burrage K, Arctander P, Mattick JS. ISIS, the intron information system, reveals the high frequency of alternative splicing in the human genome. Nat. Genet. 2000;24:340–341. doi: 10.1038/74153. [DOI] [PubMed] [Google Scholar]
- 26.Castrignano T, D'Antonio M, Anselmo A, Carrabino D, D'Onorio De Meo A, D'Erchia AM, Licciulli F, Mangiulli M, Mignone F, Pavesi G, et al. ASPicDB: a database resource for alternative splicing analysis. Bioinformatics. 2008;24:1300–1304. doi: 10.1093/bioinformatics/btn113. [DOI] [PubMed] [Google Scholar]
- 27.Raistrick CA, Day IN, Gaunt TR. Genome-wide data-mining of candidate human splice translational efficiency polymorphisms (STEPs) and an online database. PLoS One. 2010;5:e13340. doi: 10.1371/journal.pone.0013340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kahn AB, Ryan MC, Liu H, Zeeberg BR, Jamison DC, Weinstein JN. SpliceMiner: a high-throughput database implementation of the NCBI Evidence Viewer for microarray splice variant analysis. BMC Bioinformatics. 2007;8:75. doi: 10.1186/1471-2105-8-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kent WJ, Zahler AM. The intronerator: exploring introns and alternative splicing in Caenorhabditis elegans. Nucleic Acids Res. 2000;28:91–93. doi: 10.1093/nar/28.1.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pillmann H, Hatje K, Odronitz F, Hammesfahr B, Kollmar M. Predicting mutually exclusive spliced exons based on exon length, splice site and reading frame conservation, and exon sequence homology. BMC Bioinformatics. 2011;12:270. doi: 10.1186/1471-2105-12-270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nicolae M, Mangul S, Mandoiu II, Zelikovsky A. Estimation of alternative splicing isoform frequencies from RNA-Seq data. Algorithms Mol. Biol. 2011;6:9. doi: 10.1186/1748-7188-6-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Floris M, Raimondo D, Leoni G, Orsini M, Marcatili P, Tramontano A. MAISTAS: a tool for automatic structural evaluation of alternative splicing products. Bioinformatics. 2011;27:1625–1629. doi: 10.1093/bioinformatics/btr198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dimon MT, Sorber K, DeRisi JL. HMMSplicer: a tool for efficient and sensitive discovery of known and novel splice junctions in RNA-Seq data. PLoS One. 2010;5:e13875. doi: 10.1371/journal.pone.0013875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Paz I, Akerman M, Dror I, Kosti I, Mandel-Gutfreund Y. SFmap: a web server for motif analysis and prediction of splicing factor binding sites. Nucleic Acids Res. 2010;38:W281–W285. doi: 10.1093/nar/gkq444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.den Dunnen JT, Antonarakis SE. Nomenclature for the description of human sequence variations. Hum. Genet. 2001;109:121–124. doi: 10.1007/s004390100505. [DOI] [PubMed] [Google Scholar]