Abstract
Basic taste qualities like sour, salty, sweet, bitter and umami serve specific functions in identifying food components found in the diet of humans and animals, and are recognized by proteins in the oral cavity. Recognition of bitter taste and aversion to it are thought to protect the organism against the ingestion of poisonous food compounds, which are often bitter. Interestingly, bitter taste receptors are expressed not only in the mouth but also in extraoral tissues, such as the gastrointestinal tract, indicating that they may play a role in digestive and metabolic processes. BitterDB database, available at http://bitterdb.agri.huji.ac.il/bitterdb/, includes over 550 compounds that were reported to taste bitter to humans. The compounds can be searched by name, chemical structure, similarity to other bitter compounds, association with a particular human bitter taste receptor, and so on. The database also contains information on mutations in bitter taste receptors that were shown to influence receptor activation by bitter compounds. The aim of BitterDB is to facilitate studying the chemical features associated with bitterness. These studies may contribute to predicting bitterness of unknown compounds, predicting ligands for bitter receptors from different species and rational design of bitterness modulators.
INTRODUCTION
The bitterness recognition puzzle
The mammalian sense of taste is mediated by receptor proteins located in the oral cavity. Each of the five basic taste qualities – sour, salty, sweet, bitter and umami, serves a specific function in identifying components found in an animal's diet (1,2). Since many toxic plant metabolites taste bitter to mammals, bitter taste receptors are thought to have evolved to protect the organism against the ingestion of poisonous food compounds. Bitter compounds, surveyed in (3–7), are very diverse in terms of their chemical structure and physicochemical properties (8,9).
How can hundreds or more of structurally diverse compounds be detected by a limited number of receptors is an intriguing question. In particular, in humans, bitter-taste perception is mediated by 25 G-protein coupled receptors (GPCRs) of the hTAS2R gene family (10). Characterization of the receptive range of many hTAS2Rs (11,12) suggests that, this receptor family consists of both broadly tuned and narrowly tuned receptors. Apparently, individual hTAS2Rs are activated by a battery of both structurally related and unrelated bitter compounds, while preserving selectivity for chemical groups and even stereo-selectivity (11,13,14). The structural basis for hTAS2Rs’ unique ability to recognize numerous chemically diverse, and low-affinity (micromolar range) agonists is not fully understood. Assignment of bitter tastants to individual receptors has been accomplished for some ligands (11,12,14–16) but remains unknown for many others.
Not only taste perception
Recent reports about expression of taste receptors in non gustatory tissues suggest that these proteins may have additional functions apart from detection of taste (8,17,18). Of the extraoral tissues expressing taste receptors, the gastrointestinal tract has received much attention since mounting evidence indicates that after being ingested, bitter tastants might play important regulatory roles in digestive and metabolic processes (18). hTAS2Rs were also found in human airway smooth muscle tissue and the effect of bitter tastants on function of human bronchi is currently being studied (19,20).
Identification and design of modulators
Studying bitter molecules and designing selective and potent agonists and antagonists of bitter taste receptors has broad applications, both in the field of food sciences and for therapeutic indications. To date, antagonist molecules have been found by massive screening (21) and unexpectedly, when studying bitter taste receptor signaling (22). Electronic databases of compounds, such as the publicly available ZINC database (23) (http://zinc.docking.org/), have been extensively used in recent years for computer-aided drug design. The need for focused and specialized resources of compounds for virtual screening is growing as computational methods are being applied to novel research fields beyond classical drug discovery. BitterDB is an important addition to the recent databases in the field of chemical senses, such as sweet tastant molecules database (24) and database of volatile compounds relevant to odors and scents (25).
Motivation
To summarize, taste recognition is important for food sensing and possibly plays additional physiological roles. Characterizing molecules as bitter and assigning them to a specific receptor represents a crucial step towards prediction and rational modulation of bitterness. Creating a database of known bitter compounds, which, in addition to associations between bitter molecules and their receptors, also contains the chemicophysical properties of the ligands and mutagenesis data relevant for ligand binding [e.g. (26,27)], is an essential step in this direction.
DATABASE OVERVIEW
BitterDB overview/description
BitterDB currently contains more than 550 compounds that were cited in the literature as bitter. For each compound, BitterDB offers information regarding its molecular properties, references for the compound's bitterness, including additional information about the bitterness category of the compound (e.g. a ‘bitter-sweet’ or ‘slightly bitter’ annotation), different compound identifiers (smiles, CAS registry number, IUPAC systematic name), an indication whether the compound is derived from a natural source or is synthetic, a link to the compound's PubChem entry (28) and different file formats for downloading (sdf, image, smiles).
Around 100 bitter compounds have been experimentally linked to their corresponding human bitter taste receptors (12,15,16,29,30). For those compounds, BitterDB provides additional information, including links to the publications indicating these ligand–receptor interactions, the effective concentration for receptor activation and/or the EC50 value and links to the associated bitter taste receptors entries in the BitterDB.
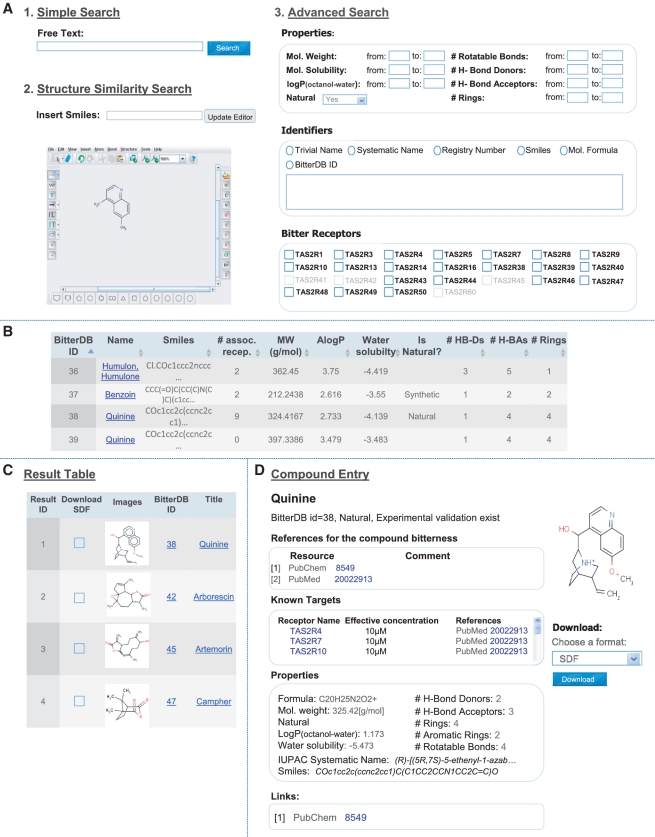
Importantly, the BitterDB database offers its users a rich interface for querying and browsing the bitter compounds dataset. BitterDB is designed to enable the user to easily find specific bitter compounds, as well as to perform more advanced searches for a range of bitter compounds with specific properties. The ‘simple search’ option allows one to retrieve bitter compounds by name, identifiers (smiles description, PubChem ID, CAS registry number) or a specific associated receptor (Figure 1A). The ‘advanced search’ allows one to retrieve compounds that fit different criteria, such as a combination of specific physical properties (see Example 1 and Figure 1A) or a combination of associated human bitter taste receptors. The 2D structure similarity search allows one to retrieve bitter compounds that are similar to a chemical structure query (Example 2 and Figure 1A). In addition to the different querying options, the user can browse through a table with all the BitterDB compounds and their properties. The compounds in the table can be sorted according to different criteria (Example 3 and Figure 1B).
Figure 1.
Screenshots of the BitterDB interface. (A) Illustration of three available search options: (i) Simple search: search compounds using keywords. (ii) Structure-based similarity search: 2D similarity search using different structural formats (drawing, smiles and user uploaded structure files). (iii) Advanced search, including searching by chemical descriptors, molecule identifiers and by associated human bitter taste receptors. Orphan receptors (shown in gray) cannot be searched by ligands. (B) The BitterDB compound browse table (which can be sorted by different criteria). (C) An example of a query result page: the user can download an SDF file of selected compounds from the result page. (D) A representative compound entry page.
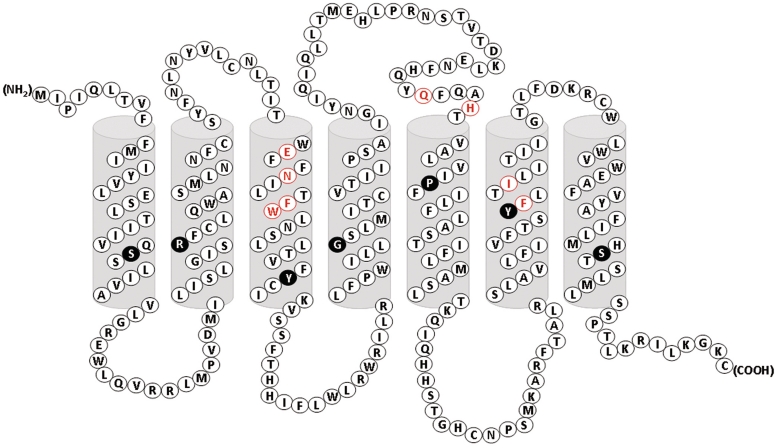
BitterDB also contains data about the 25 known human bitter taste receptors. Several properties are displayed for each receptor: molecular weight, protein sequence, genomic location [according to the UCSC genome browser (31)], as well as links to the corresponding UCSC Genome browser (31) and UniProt (32) entries. For each receptor, a list of its known bitter ligands, collected from the literature, is displayed with links to the individual ligand entries and an option to download ligand structures as a SDF file. A 2D protein diagram, which offers an intuitive way of looking at the sequence of the receptor and its trans-membrane (TM) regions, is displayed for each receptor. For several receptors, data regarding mutagenesis experiments performed on the receptor sequence were found in the literature. For these receptors we generated a mutation table that summarizes the mutagenesis experiments and their effects on receptor function.
The bitter taste receptors can be searched using different criteria: name, known ligands and UniProt accession number. Using a table that presents all the bitter taste receptors and some information about them, the user can browse and also sort by various options, such as the number of bitter ligands associated with the receptor.
In addition, BitterDB offers two extra features: a local BLAST service to determine the local similarity between a query sequence and the different human bitter taste receptors (Example 4), and a global alignment of the 25 human bitter taste receptors. The alignment was generated using ClustalW2 (33), and displays the secondary structure of each receptor, as predicted by the TOPCONS server (34). In addition, the most conserved residue in each TM helix is annotated by its Ballesteros–Weinstein numbering scheme (35); the most conserved residue in a given TM is assigned the index X.50, where X is the TM number, and the remaining residues are numbered relative to this position.
Importantly, BitterDB offers an ‘upload’ option, by which the users are encouraged to submit information about bitter compounds. These data will be reviewed by the authors and uploaded to the database accordingly. The users can join the BitterDB mailing list in order to receive database updates.
MATERIALS AND METHODS
Data acquisition
Data were collected from the literature and from various web resources. Chemical compounds were included in the database only if they were described as bitter in the literature, or if they were reported experimentally as capable of activating at least one human bitter taste receptor. The main resources were (numbers in brackets indicate the numbers of compounds retrieved): the Merck index (36) [300], Fenaroli's Handbook of Flavor Ingredients (37) [94] and a recent publication by Meyerhof et al. (12) [98]. Most of the 2D chemical structures of the compounds and the IUPAC systematic names were taken from PubChem (28). A set of physical descriptors such as molecular weight, logP, molecular solubility, water solubility, H-bond donors, H-bond acceptors and the number of rings, aromatic rings and rotatable bonds were computed for each compound using the ‘calculate molecular properties protocol’ in Discovery Studio 2.5.5 (DS2.5.5; Accelrys, Inc.).
Data on human bitter taste receptors were taken mainly from the UniprotKB database (32). Experimental mutagenesis data on the bitter taste receptors were collected from the literature. The main resource was Brockhoff et al. (26). The secondary structure/topology of the receptors was predicted by the TOPCONS server (34).
Structure
Similarity searches
MarvinSketch, 5.3.8, 2010 (ChemAxon, Inc.), an advanced chemical editor for drawing chemical structures by the user, was implemented as the built-in molecular editor. The structure similarity search was implemented with Open Babel Package, version 2.2.3 http://openbabel.sourceforge.net/ (accessed December 2010). The degree of similarity between the query molecule and database entries is assessed by the Tanimoto coefficient using the MACCS fingerprints. MACCS fingerprints are a set of bitstrings; each bit refers to the presence or absence of one of the 166 MACCS keys in a given molecule. The MACCS fingerprints were pre-calculated for all the BitterDB molecules and are calculated during the search for the query structure, in order to compare it to the database entries. The Tanimoto coefficient, which is a number between 0 and 1 (1 corresponding to ‘maximum similarity’), is used to measure the distance between two molecules (query and database entry). The Tanimoto coefficient calculates the number of bit positions set to 1 in both fingerprints, divided by the number of bit positions set to 1 in at least one of the fingerprints. The top 20 results are returned.
Blast
BitterDB contains a local BLAST service, powered by the NCBI BLAST package (38). We created a custom database of the 25 protein sequences of the human bitter taste receptors whose query sequence is searched against. The user is able to control the different search parameters, such as scoring matrix, gap cost and others.
Visualization tools/features
BitterDB contains a range of visualization tools. These include a 2D compound structure display and a 2D image for each compound, which are generated using Marvin, 5.3.8, 2010 (ChemAxon http://www.chemaxon.com/), a 2D transmembrane protein diagram with indication of the most conserved residue in each helix and residues for which mutation data is available (see Figure 2) for each human bitter receptor, which is based on the TOPCONS secondary structure prediction (34) and an annotated bitter receptor alignment, which is powered by JalView (39).
Figure 2.
2D transmembrane protein diagram. A convenient representation of the receptor sequence and its TM regions, shown here for hTAS2R16. The seven TM helices are displayed as predicted by Topcons. The most conserved residue in each helix X (BW number X.50) has black background. Residues for which mutation data are available are marked with red, and information about the mutations can be found in the site ‘Known Mutations Table’ section below the diagram http://bitterdb.agri.huji.ac.il/bitterdb/Receptor.php?id=16#ReceptorKnownMutations.
Web Server
BitterDB was designed as a relational database on a MariaDB server http://mariadb.org/. The website was built with the PHP server side, and web access is enabled via an Apache HTTP Server. The site is best viewed in Firefox, Opera, Chrome and Safari browsers.
EXAMPLES
Examples of BitterDB usage are provided below. A movie illustrating each example was recorded from the BitterDB website; the movies can be accessed from the ‘Examples’ link of the database, http://bitterdb.agri.huji.ac.il/bitterdb/examples.php
Possible usage: filtering molecules using their properties
Specific Example #1: Searching for bitter molecules that satisfy Lipinski's rule of five criteria
It is often useful to obtain a subset of molecules having particular physicochemical properties. As an example, we demonstrate how one can obtain a subset of bitter molecules that satisfy Lipinski's rule of five (40). This rule, based on properties of known orally available drugs, states that in general, an orally active drug has no more than one violation of the following criteria: Not more than 5 H-bond donors or 10 H-bond acceptors, a molecular mass under 500Da, and an octanol-water partition coefficient (logP) of less than five (40). In movie #1 (http://bitterdb.agri.huji.ac.il/bitterdb/examples.php#ex1), we demonstrate how, using Lipinksi's criteria as a filter, we retrieve 83% of the molecules in our database. These molecules satisfy Lipniski's rule of five and therefore can be considered ‘drug-like’.
Possible usage: finding bitter compounds using the 2D similarity search
Specific Example #2: Searching for compounds with a 2D structure similarity to aristolochic acid
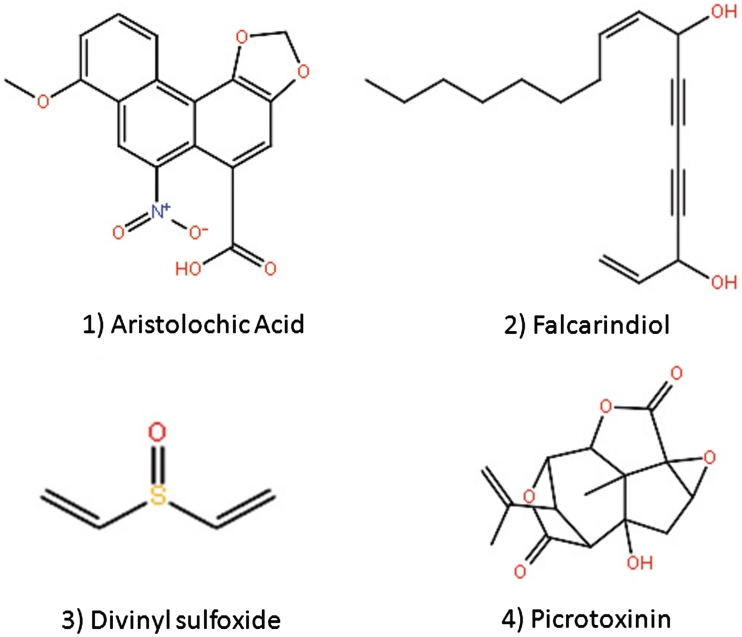
It is commonly assumed that chemically similar compounds may bind to the same receptor (41–43). However, the chemical features of ligands of the same bitter taste receptor are very diverse (see Figure 3). It is therefore worthwhile to extend our exploration of the chemical space around known ligands of bitter taste receptors, in order to gain more information about the physicochemical determinants for bitterness. Furthermore, since most of the bitter compounds are not yet assigned to a particular bitter taste receptor target, similarity of a query compound to a ligand with known receptor association(s) may provide a helpful clue.
Figure 3.
A subset of four hTAS214 ligands demonstrating a high degree of structural variability. These four structurally diverse compounds, which are all hTAS214 ligands, demonstrate that hTAS214 ligands lie in a broad chemical spectrum that includes compounds from different classes such as aromatic polycyclic compounds (1), polyacetylenes (2), sulfoxides (3) and cycloparaffins (4).
In Example 2 (http://bitterdb.agri.huji.ac.il/bitterdb/examples.php#ex2) we searched for compounds with a 2D structure similarity to aristolochic acid, a known ligand of the bitter taste receptor hTAS2R14 (as well as hTAS2R43 and hTAS2R44) (12). Nine of the top ten results were not yet assigned to any bitter receptor target. However, result number six, Noscapine, is also a ligand of hTAS2R14. This interesting result suggests that additional aromatic compounds that bear resemblance to aristolochic acid could be potential hTAS2R14 ligands.
Possible usage: Browsing and sorting bitter compounds
Specific Example #3: Sorting compounds by their logP value
Although the structural and physicochemical determinants for bitterness remain largely unknown, there are some chemical properties within specific chemical families that are related to bitterness (44–46). For example, several studies proposed a correlation between hydrophobicity and bitterness (44,45). Additionally, a recent study showed that bitterness of the soy peptides is predominantly associated with molecular mass (47).
Hence, it is interesting to sort the dataset of BitterDB compounds by different molecular properties and to explore the associated ranges of bitterness. In this example (http://bitterdb.agri.huji.ac.il/bitterdb/examples.php#ex3), we sorted the dataset according to the logP-value, which is one of the means of measuring small molecules’ hydrophobicity.
Possible usage: relating bitter taste receptors from different species to human bitter receptors
Specific Example #4: finding human bitter taste receptors with sequence similarity to the zebra fish (Danio rerio) bitter taste receptor TAS2R5
Organisms’ sense of bitter taste helps them avoid toxic and harmful substances. Although research on bitter taste in humans has advanced rapidly, less is known about taste perception in other organisms. Thus, it is beneficial to relate novel taste receptors to human ones, and learn about potential common ligands, relevant known mutations, etc.
The Zebrafish has only four intact TAS2R genes as opposed to 25 genes in humans. In this example (http://bitterdb.agri.huji.ac.il/bitterdb/examples.php#ex4), the zebrafish TAS2R5 receptor protein sequence was searched against the human bitter taste receptors sequences in BitterDB. The goal was to find the most similar human receptors using the BitterDB local BLAST service. The human receptor with the highest score for local similarity is hTAS2R20 (also known as hTAS2R49). hTAS2R20 has two known ligands—diphenidol and cromolyn, which, to the best of our knowledge, have not yet been tested on zebrafish T2Rs. The second most similar receptor, hTAS2R42, is still an orphan receptor, and the next one, hTAS2R4, has 15 known ligands. Interestingly, one of the hTAS2R4 ligands is denatonium benzoate, which was also reported as a zebrafish TAS2R5 ligand (48). Detailed studies of ligand–receptor interactions from different species are needed to better understand the range of activity of the different receptors and its relation to the number of receptor types in a given species. Yet, as a first guess, the receptor similarity option can be used to provide a preliminary list of suspected ligands, based on the ligands of human receptors with the highest sequence similarity, which were already de-orphanized.
SUMMARY AND OUTLOOK
Our goals in the present study were (i) to compile a comprehensive dataset of all chemical structures that have been cited in the literature as having bitter taste, and (ii) to assign bitter ligands to their cognate receptors in humans, where possible. To the best of our knowledge, although a proprietary database of bitter compounds was established in the past (5), BitterDB is the first publicly available, electronically searchable database of bitter molecules.
Interestingly, not only does the receptor receptive range vary within bitter taste receptors (12)—also the number of functional bitter taste receptors varies among different species [e.g. (49)]. The number of receptors in a given species may relate to the receptors’ receptive range, the effects of variation within the population (4) and the environmental pressure. The ability to understand and predict these effects relies on the knowledge of what is ‘bitter’ (or aversive) to different species, and on the ability to cluster ligands based on their chemical similarity. BitterDB will be extended in the future to include additional species, and thus will contribute to elucidating evolutionary aspects of bitter taste perception.
Fundamental questions as well as potential therapeutic applications of the modulation of bitter taste receptors continue to motivate further development of the BitterDB database. Future extensions may include implementing 3D similarity searches of compounds, modeling 3D structures of the receptors and mapping single nucleotide polymorphisms (SNPs) onto these models.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Files.
FUNDING
Niedersachsen-Israel Research cooperation fund and BSF grant number 2007296. Funding for open access charge: Intramural grant from the Hebrew University.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors are grateful to Dvorah Art-Weisman, Tomer Levy and Marylin Safran for technical help and Dr Merav Fichman for discussions. MYN holds the Women's League for Israel Senior Lectureship for Nutrition.
REFERENCES
- 1.Chandrashekar J, Hoon MA, Ryba NJ, Zuker CS. The receptors and cells for mammalian taste. Nature. 2006;444:288–294. doi: 10.1038/nature05401. [DOI] [PubMed] [Google Scholar]
- 2.Chaudhari N, Roper SD. The cell biology of taste. J. Cell. Biol. 2010;190:285–296. doi: 10.1083/jcb.201003144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Drewnowski A, Gomez-Carneros C. Bitter taste, phytonutrients, and the consumer: a review. Am J. Clin. Nutr. 2000;72:1424–1435. doi: 10.1093/ajcn/72.6.1424. [DOI] [PubMed] [Google Scholar]
- 4.Hofmann T. Identification of the key bitter compounds in our daily diet is a prerequisite for the understanding of the hTAS2R gene polymorphisms affecting food choice. Ann N Y Acad. Sci. 2009;1170:116–125. doi: 10.1111/j.1749-6632.2009.03914.x. [DOI] [PubMed] [Google Scholar]
- 5.Rodgers S, Busch J, Peters H, Christ-Hazelhof E. Building a tree of knowledge: analysis of bitter molecules. Chem. Senses. 2005;30:547–557. doi: 10.1093/chemse/bji048. [DOI] [PubMed] [Google Scholar]
- 6.Rodgers S, Glen RC, Bender A. Characterizing bitterness: identification of key structural features and development of a classification model. J. Chem. Inf. Model. 2006;46:569–576. doi: 10.1021/ci0504418. [DOI] [PubMed] [Google Scholar]
- 7.Maehashi K, Huang L. Bitter peptides and bitter taste receptors. Cell Mol. Life Sci. 2009;66:1661–1671. doi: 10.1007/s00018-009-8755-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Behrens M, Meyerhof W. Bitter taste receptors and human bitter taste perception. Cell Mol. Life Sci. 2006;63:1501–1509. doi: 10.1007/s00018-006-6113-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Meyerhof W, Born S, Brockhoff A, Behrens M. Molecular biology of mammalian bitter taste receptors. A review. Flavour Frag. J. 2011;26:260–268. [Google Scholar]
- 10.Behrens M, Meyerhof W. Mammalian bitter taste perception. Results Probl. Cell. Differ. 2009;47:203–220. doi: 10.1007/400_2008_5. [DOI] [PubMed] [Google Scholar]
- 11.Brockhoff A, Behrens M, Massarotti A, Appendino G, Meyerhof W. Broad tuning of the human bitter taste receptor hTAS2R46 to various sesquiterpene lactones, clerodane and labdane diterpenoids, strychnine, and denatonium. J. Agric. Food Chem. 2007;55:6236–6243. doi: 10.1021/jf070503p. [DOI] [PubMed] [Google Scholar]
- 12.Meyerhof W, Batram C, Kuhn C, Brockhoff A, Chudoba E, Bufe B, Appendino G, Behrens M. The molecular receptive ranges of human TAS2R bitter taste receptors. Chem. Senses. 2010;35:157–170. doi: 10.1093/chemse/bjp092. [DOI] [PubMed] [Google Scholar]
- 13.Bufe B, Breslin PA, Kuhn C, Reed DR, Tharp CD, Slack JP, Kim UK, Drayna D, Meyerhof W. The molecular basis of individual differences in phenylthiocarbamide and propylthiouracil bitterness perception. Curr. Biol. 2005;15:322–327. doi: 10.1016/j.cub.2005.01.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bufe B, Hofmann T, Krautwurst D, Raguse JD, Meyerhof W. The human TAS2R16 receptor mediates bitter taste in response to beta-glucopyranosides. Nat. Genet. 2002;32:397–401. doi: 10.1038/ng1014. [DOI] [PubMed] [Google Scholar]
- 15.Sainz E, Cavenagh MM, Gutierrez J, Battey JF, Northup JK, Sullivan SL. Functional characterization of human bitter taste receptors. Biochem. J. 2007;403:537–543. doi: 10.1042/BJ20061744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sakurai T, Misaka T, Ishiguro M, Masuda K, Sugawara T, Ito K, Kobayashi T, Matsuo S, Ishimaru Y, Asakura T, et al. Characterization of the beta-D-glucopyranoside binding site of the human bitter taste receptor hTAS2R16. J. Biol. Chem. 2010;285:28373–28378. doi: 10.1074/jbc.M110.144444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kinnamon SC. Taste receptor signalling - from tongues to lungs. Acta Physiol. 2011 doi: 10.1111/j.1748-1716.2011.02308.x. doi:10.1111/j.1748-1716.2011.02308.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Behrens M, Meyerhof W. Gustatory and extragustatory functions of mammalian taste receptors. Physiol. Behav. 2011 doi: 10.1016/j.physbeh.2011.02.010. doi:10.1016/j.physbeh.2011.02.010. [DOI] [PubMed] [Google Scholar]
- 19.Deshpande DA, Wang WC, McIlmoyle EL, Robinett KS, Schillinger RM, An SS, Sham JS, Liggett SB. Bitter taste receptors on airway smooth muscle bronchodilate by localized calcium signaling and reverse obstruction. Nat. Med. 2011;16:1299–1304. doi: 10.1038/nm.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morice AH, Bennett RT, Chaudhry MA, Cowen ME, Griffin SC, Loubani M. Effect of bitter tastants on human bronchi. Nat. Med. 2011;17:775. doi: 10.1038/nm0711-775. [DOI] [PubMed] [Google Scholar]
- 21.Slack JP, Brockhoff A, Batram C, Menzel S, Sonnabend C, Born S, Galindo MM, Kohl S, Thalmann S, Ostopovici-Halip L, et al. Modulation of bitter taste perception by a small molecule hTAS2R antagonist. Curr. Biol. 2010;20:1104–1109. doi: 10.1016/j.cub.2010.04.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Greene TA, Alarcon S, Thomas A, Berdougo E, Doranz BJ, Breslin PA, Rucker JB. Probenecid inhibits the human bitter taste receptor TAS2R16 and suppresses bitter perception of salicin. PLoS One. 6:e20123. doi: 10.1371/journal.pone.0020123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Irwin JJ, Shoichet BK. ZINC–a free database of commercially available compounds for virtual screening. J. Chem. Inf. Model. 2005;45:177–182. doi: 10.1021/ci049714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ahmed J, Preissner S, Dunkel M, Worth CL, Eckert A, Preissner R. SuperSweet–a resource on natural and artificial sweetening agents. Nucleic Acids Res. 2011;39:D377–D382. doi: 10.1093/nar/gkq917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dunkel M, Schmidt U, Struck S, Berger L, Gruening B, Hossbach J, Jaeger IS, Effmert U, Piechulla B, Eriksson R, et al. SuperScent–a database of flavors and scents. Nucleic Acids Res. 2009;37:D291–D294. doi: 10.1093/nar/gkn695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brockhoff A, Behrens M, Niv MY, Meyerhof W. Structural requirements of bitter taste receptor activation. Proc. Natl Acad. Sci. USA. 2010;107:11110–11115. doi: 10.1073/pnas.0913862107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Levit A, Barak D, Behrens M, Meyerhof W, Niv MY. Homology model-assisted elucidation of binding sites in GPCRs. Methods Mol. Biol. 2011 doi: 10.1007/978-1-62703-023-6_11. Accepted for publication. [DOI] [PubMed] [Google Scholar]
- 28.Bolton E, Wang Y, Thiessen P, Bryant S. Annual Reports in Computational Chemistry. Vol. 4. Washington, DC: American Chemical Society; 2008. [Google Scholar]
- 29.Intelmann D, Batram C, Kuhn C, Haseleu G, Meyerhof W, Hofmann T. Three TAS2R bitter taste receptors mediate the psychophysical responses to bitter compounds of hops (Humulus lupulus L.) and beer. Chemosensory Perception. 2009;2:118–132. [Google Scholar]
- 30.Dotson CD, Zhang L, Xu H, Shin YK, Vigues S, Ott SH, Elson AE, Choi HJ, Shaw H, Egan JM, et al. Bitter taste receptors influence glucose homeostasis. PLoS One. 2008;3:e3974. doi: 10.1371/journal.pone.0003974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Magrane M, Consortium U. UniProt Knowledgebase: a hub of integrated protein data. Database. 2011;2011 doi: 10.1093/database/bar009. bar009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ogden TH, Rosenberg MS. Alignment and topological accuracy of the direct optimization approach via POY and traditional phylogenetics via ClustalW + PAUP*. Syst. Biol. 2007;56:182–193. doi: 10.1080/10635150701281102. [DOI] [PubMed] [Google Scholar]
- 34.Hennerdal A, Elofsson A. Rapid membrane protein topology prediction. Bioinformatics. 2011;27:1322–1323. doi: 10.1093/bioinformatics/btr119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ballesteros JA, Weinstein H. Integrated methods for the construction of three dimensional models and computational probing of structure function relations in G protein-coupled receptors. Methods Neurosci. 1995;5:366–428. [Google Scholar]
- 36. O'Neil,M.J., Heckelman,P.E., Koch,C.B., Roman,K.J. (eds). (2006). Fourteenth Edition ed. Merck & Co., Inc., Whitehouse Station, NJ, USA.
- 37. Burdock,G.A. (2005) Fenaroli's handbook of flavor ingredients. 5th edn. CRC Press, Boca Raton, Fla.
- 38.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 39.Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ. Jalview Version 2–a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lipinski CA, Lombardo F, Dominy BW, Feeney PJ. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 2001;46:3–26. doi: 10.1016/s0169-409x(00)00129-0. [DOI] [PubMed] [Google Scholar]
- 41.Allen JA, Roth BL. Strategies to discover unexpected targets for drugs active at g protein-coupled receptors. Annu. Rev. Pharmacol. Toxicol. 2011;51:117–144. doi: 10.1146/annurev-pharmtox-010510-100553. [DOI] [PubMed] [Google Scholar]
- 42.Keiser MJ, Irwin JJ, Shoichet BK. The chemical basis of pharmacology. Biochemistry. 2010;49:10267–10276. doi: 10.1021/bi101540g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Keiser MJ, Setola V, Irwin JJ, Laggner C, Abbas AI, Hufeisen SJ, Jensen NH, Kuijer MB, Matos RC, Tran TB, et al. Predicting new molecular targets for known drugs. Nature. 2009;462:175–181. doi: 10.1038/nature08506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ney KH. Prediction of bitterness of peptides from their amino acid composition. Z. Lebensm.-Untersuch. Forch. 1971;147:64–68. [Google Scholar]
- 45.Cravotto G, Nano GM, Binello A, Spagliardi P, Seu G. Chemical and biological modification of cynaropicrin and grosheimin: a structure–bitterness relationship study. J. Sci. Food Agric. 2005;85:1757–1764. [Google Scholar]
- 46.Kim MR, Yukio K, Kim KM, Lee CH. Tastes and structures of bitter peptide, asparagine-alanine-leucine-proline-glutamate, and its synthetic analogues. J. Agric. Food Chem. 2008;56:5852–5858. doi: 10.1021/jf7036664. [DOI] [PubMed] [Google Scholar]
- 47.Cho MJ, Unklesbay N, Hsieh FH, Clarke AD. Hydrophobicity of bitter peptides from soy protein hydrolysates. J. Agric. Food Chem. 2004;52:5895–5901. doi: 10.1021/jf0495035. [DOI] [PubMed] [Google Scholar]
- 48.Oike H, Nagai T, Furuyama A, Okada S, Aihara Y, Ishimaru Y, Marui T, Matsumoto I, Misaka T, Abe K. Characterization of ligands for fish taste receptors. J. Neurosci. 2007;27:5584–5592. doi: 10.1523/JNEUROSCI.0651-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Davis JK, Lowman JJ, Thomas PJ, ten Hallers BF, Koriabine M, Huynh LY, Maney DL, de Jong PJ, Martin CL, Thomas JW. Evolution of a bitter taste receptor gene cluster in a New World sparrow. Genome Biol. Evol. 2010;2:358–370. doi: 10.1093/gbe/evq027. [DOI] [PMC free article] [PubMed] [Google Scholar]