Abstract
Genome sequencing has been democratized by second-generation technologies, and even small labs can sequence metazoan genomes now. In this article, we describe ‘959 Nematode Genomes’—a community-curated semantic wiki to coordinate the sequencing efforts of individual labs to collectively sequence 959 genomes spanning the phylum Nematoda. The main goal of the wiki is to track sequencing projects that have been proposed, are in progress, or have been completed. Wiki pages for species and strains are linked to pages for people and organizations, using machine- and human-readable metadata that users can query to see the status of their favourite worm. The site is based on the same platform that runs Wikipedia, with semantic extensions that allow the underlying taxonomy and data storage models to be maintained and updated with ease compared with a conventional database-driven web site. The wiki also provides a way to track and share preliminary data if those data are not polished enough to be submitted to the official sequence repositories. In just over a year, this wiki has already fostered new international collaborations and attracted newcomers to the enthusiastic community of nematode genomicists. www.nematodegenomes.org.
INTRODUCTION
The nematode Caenorhabditis elegans was the first animal to have its genome completely sequenced in 1998 (1). Since then, second-generation sequencing technologies have revolutionized and democratized the field of genome sequencing. Even small labs can now sequence their favourite nematodes in a few weeks for a few thousand dollars. By 2012, we anticipate that more than 100 nematode genomes will be sequenced, a happy state of affairs for those of us who study this most abundant and diverse Metazoan phylum.
The only problem with rapid and inexpensive sequencing is that it is becoming harder to keep track of which genomes are being sequenced, who is sequencing them, what stage the genome projects are at, and where one can get early access to the data. The nucleotide sequence archives (GenBank/EMBL/DDBJ) (2) are the de facto storehouses for complete and published genomes. However, as the bottleneck of a genome project has shifted from sequencing to analysis, which can take months, it has become imperative to have a place to share information about the project before it is published. Inspired by ArthropodBase (www.arthropodgenomes.org), the 959 Nematode Genomes (959NG) wiki was created in early 2010 to meet this need and can be accessed at www.nematodegenomes.org.
959NG is unlike existing genome and transcriptome database web sites such as WormBase (3) and NemBase (4) because, instead of storing the relationships between genes, proteins and DNA sequences, it stores the relationships between people, institutions and sequencing projects at various stages of completion. The goal is to connect users, and make it easy for them to form collaborations and share data. The platform choice reflects this goal as we describe in the ‘Software’ section.
Why (Only) 959NG?
Unlike the 1000 Human Genomes (www.1000genomes.org) or Genome 10 K (genome10k.soe.ucsc.edu) sequencing projects, the effort to sequence as many nematodes as possible is a distributed, bottom-up enterprise. We picked 959 as an initial target because all adult female hermaphrodite C. elegans have exactly 959 somatic cells. The definition of the embryonic lineage of C. elegans from fertilized zygote to fertile adult was a milestone in C. elegans developmental biology. Just as the tree of the C. elegans embryonic lineage was a key underpinning of later work on this model nematode, we hope that a nematode phylogeny with 959 genome-sequenced taxa will underpin the investigation of nematode biology in general. Obviously, we do not limit the vision to these few genomes: with 23 000 species described, and an estimated 1–2 million species undescribed, the scope for genomic exploration of Nematoda is vast.
FEATURES
959NG is a wiki and thus very easy for end-users to edit and interact with. As it is based on the Semantic MediaWiki (SMW) platform, it also allows pages to store properties and relationships to other pages. These properties and relationships can be queried by anyone.
Editable Taxonomy
We offer a view of the taxonomy of the phylum Nematoda, pre-loaded with all species that have data present in EMBL/GenBank/DDBJ. Clicking on any node in the taxonomic tree of nematodes shows the sequencing status of all species below that taxon. Each node also provides links to the NCBI page for that taxon and the Expressed Sequence Tags (ESTs) available for any species within that taxon (Figure 1). The initial tree was populated using the NCBI taxonomy (www.ncbi.nlm.nih.gov/taxonomy) but the more widely used Blaxter clades (5) and Helder clades (6) were easy to incorporate into the tree because of the SMW architecture. Users can add new species. See the ‘Software’ section for more details.
Figure 1.
Systematic tree of Nematoda, with a few taxonomic nodes expanded to show how the Blaxter and Helder classifications were incorporated into the tree.
Species and Strain Information
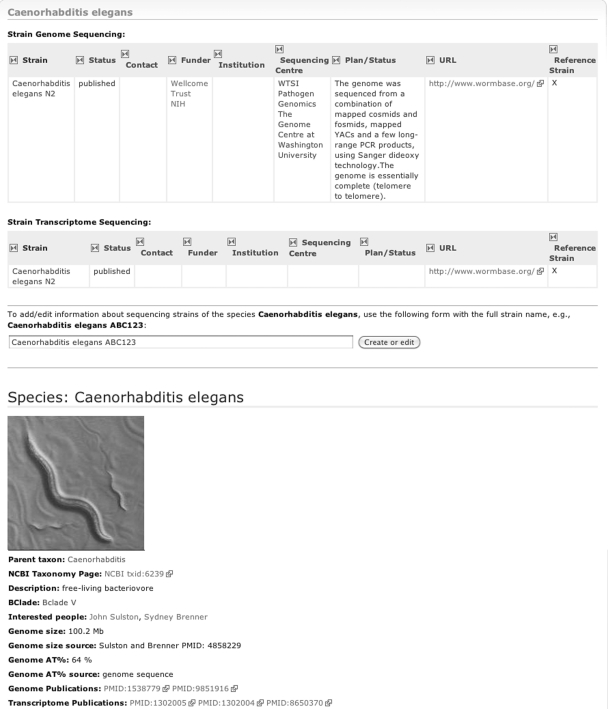
For each species, several pieces of information are stored and displayed, such as a short description, its NCBI taxonomic identifier, a picture, as well as some facts about genome size and nucleotide frequency, if known. Species pages also store names of people interested in that species. Each species can have one or more strains with a genome and transcriptome sequencing status that includes links to the funding bodies and the sequencing centres contributing to the sequencing projects (Figure 2).
Figure 2.
Species page for C. elegans displaying information for the species as well as the status of strains that have been sequenced.
All page properties are stored internally as Resource Description Framework (RDF) triples which are expressions with three parts: subject, predicate and object. An example of an RDF triple is ‘Brugia malayi TRS: Strain genome status: Published’. Although some properties are integer or text values, other properties define relationships to pages, such as ‘Trichinella spiralis: Has interested party: Makedonka Mitreva’ which links to a person page.
Persons and Organizations
Because the main goal of 959NG is to connect users, people and organization pages are as important as species pages. These pages store personal and institutional URLs, contact information as well as relationships to the species such as ‘is genome contact for’ and ‘is interested in species’.
Queries
SMW sites allow users to add new properties that the original web site creators may not have thought of. These properties and relationships can be queried to generate useful dynamic tables. Using the species, strain, people and organization properties, any user can create queries to collate and display information. The following queries are already implemented and linked to from the home page as potentially useful starting points:
species with published genomes;
species with genomes being sequenced; and
species for which sequencing has been proposed.
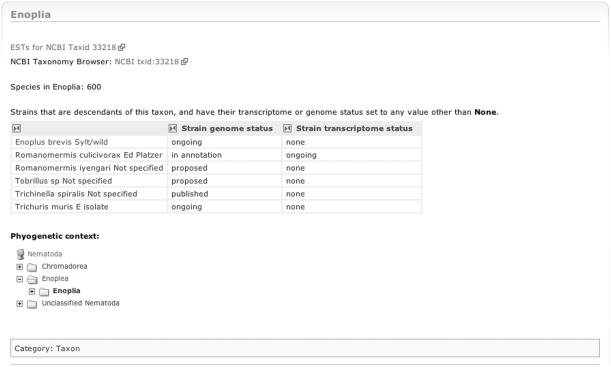
In addition, clicking on a node in the taxonomic tree displays the result of the query ‘Species under this taxon that have their sequencing status set to anything other than “None”’ (Figure 3).
Figure 3.
Page for the taxonomic node Enoplia, showing NCBI Taxonomy and NCBI EST link-outs, as well as the results of the query ‘Species and strains under this taxon with their sequencing status set to anything other than “None”’.
New queries and information mash-ups can be added by users on any page if they know the SMW query syntax. For example, the following queries are trivial to run from the ‘Semantic Search’ page:
List of strains sequenced by the funding body NIH: [[Strain_genome_funder::NIH]]
Species in Blaxter clade III with Adenine-Thymine content greater than 70%: [[Category:Species]] [[Species_genome_at:: >70]] [[Species_bclade::Bclade_III]]
All the pages and the relationships in 959NG can also be exported in XML and RDF format, respectively, using the Special:Export and Special:ExportRDF sections of the web site.
Blast Server For Genomes in Progress
One of the most used features of 959NG is the BLAST (7) server for intermediate genome assemblies. Although generating sequence data is no longer the bottleneck in a sequencing project, quality checks, assembly, annotation and analysis of the data can take several months. The 959NG BLAST server provides a place to park intermediate data so that interested researchers can start looking for their genes or features of interest and speed up the process of research, especially in time-critical areas such as drug–target and vaccine–candidate discovery. Completed genomes will be submitted to centralized repositories (GenBank/EMBL/DDBJ) and to specialized databases such as WormBase, at which point the intermediate assemblies can be removed from the 959NG BLAST server.
SOFTWARE
SMW (semantic-mediawiki.org) is an extension to the popular MediaWiki (mediawiki.org) platform that powers Wikipedia. We chose it for the 959NG web site because (i) users are familiar with wikis and comfortable with creating and editing pages and (ii) we were not sure at the outset about the information we wanted to capture for each species and its genome sequencing status. As we show in this section, SMWs are better than traditional databases when the data model may change.
SMW Concepts
The initial setup requires an understanding of the following SMW concepts:
Categories: all pages on the site are in one of the following Categories: (i) Genome Sequencing Centre, (ii) Person, (iii), Species, (iv) Strain and (v) Taxon. A category would typically correspond to a table in a relational database.
Forms: each category normally has a specialized form to enter information for that type of page. For example, a Taxon form will have fields for ‘NCBI taxon id’ and ‘Taxon parent’, which are specific to Taxon pages.
Pages and Properties: a page is analogous to an object in a database or a row in a database table. Page properties in SMW are conceptually equivalent to object values or to columns in a database table.
Templates: templates display information about a page or a property. Each category will usually have a template that determines how the information for those types of pages should be displayed. Templates also transform values into displays. For example, the ‘PubmedID Linkout’ template takes a PubMed ID such as 20980554 and displays a URL to that article on PubMed.
Advantages of SMW
Traditional database-driven web sites have fixed data models that are defined by the developers, and end-users typically only add data within the existing framework to such web sites. One of the main advantages of our SMW site is that, as sequencing technologies and needs change, even end-users can change the types of data stored for each entity (species, person, organization, etc.). For example, when we started the web site in early 2010, we did not have strain-specific pages because only one strain was sequenced per species. However, with sequencing becoming more accessible, different strains are now being sequenced for the same species, so we used the web interface to add a new ‘Strain’ category, created a new template and a new form for strains, and thus changed the fundamental data model without once touching a database table.
The taxonomy tree is another example of how an end-user can change the data hierarchy without knowing anything about how the back-end is implemented. On our site, each taxon is a wiki page with a ‘Taxon parent’ property pointing to another taxon page, and the tree is generated dynamically based on this single property. Therefore, all we had to do to include additional sub-classifications such as Blaxter clades and Helder clades was to edit a few high-level taxon pages so that their ‘Taxon parent’ properties pointed to a new Blaxter or Helder clade page.
Another advantage of SMW is that it sits on the MediaWiki platform, which is a mature and scalable engine for serving high-capacity web sites and has a large developer base. Setting up the initial web site took only three person-days, thanks to the examples of templates and forms on another similar site (arthropodgenomes.org). The BioDBCore description of the wiki is provided in the Supplementary Data section.
FUTURE DIRECTIONS
As more genomes are sequenced and the 959NG site grows, we hope that the evolving data model for nematode genome sequencing projects will also inform other genome sequencing efforts. Most genomes these days are not finished, but are published as high-quality draft sequences, so we will need to not only store the Minimum Information about a Genome Sequence (gensc.org) and Minimum Information about a high-throughput SeQuencing Experiment (www.mged.org/minseqe), but also additional values such as CEGMA scores (8) to measure how complete the genome is. We will also develop descriptors for genome-scale genetic mapping data, derived from technologies such as restriction-site-associated DNA sequencing (RADSeq) (9), genotyping by sequencing (GBS) (10) and other methods (11), across many strains or isolates of a species.
Currently, site visitors can interrogate intermediate draft assemblies of genomes in progress only through the BLAST server. In addition, we would like to provide a basic, automatic annotation service for these incomplete genomes using RNASeq alignments and gene predictors.
CONCLUSIONS
The 959 Nematode Genomes wiki has already inspired international collaborations to sequence, annotate and interpret the genomes of key species. We know of two cases where groups who did not know of each other's efforts are now merging expertise and effort in a unified project. As additional genomes are proposed, new collaborations can be forged and cross-species analyses coordinated. We also hope that the existence of the wiki, and the enthusiastic community behind it, will serve to attract new researchers into this field. As nematode genomics moves into population genomics, this register of strains and sources will become ever more useful. SMW technology builds a system that is easy to navigate, easy to edit and, importantly, easy to develop as needs, knowledge and possibilities change.
Genomics research on nematodes (particularly C. elegans) has already delivered important information on core biological processes. Adding additional nematode genomes will allow the specific instance of C. elegans to be contextualized, and will, we hope, feed research on comparative genomics of nematodes, the evolutionary biology of genome change, and the biology of (many) parasitic nematodes, among other fields. We hope 959NG will become a one-stop site in which to forge collaborations, learn about best practice in assembly and annotation, record insights and advances and explore the genomic diversity of Nematoda.
FUNDING
This work was supported by the School of Biological Sciences at the University of Edinburgh. Funding for open access charge: Natural Environment Research Council (NERC).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We would like to thank Dan Lawson at EBI for inspiring us with his SMW site ArthropodBase and allowing us to use his templates and forms as a starting point. Dan Bolser at the University of Dundee, Yaron Koren of WikiWorks and the rest of the SMW community were very helpful and patiently answered questions on online forums. The University of Edinburgh provides hosting space for nematodegenomes.org.
REFERENCES
- 1.C elegans Genome Sequencing Consortium. Genome sequence of the nematode C. elegans: A platform for investigating biology. Science. 1998;282:2012–2018. doi: 10.1126/science.282.5396.2012. [DOI] [PubMed] [Google Scholar]
- 2.Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW. GenBank. Nucleic Acids Res. 2011;39:D32–D37. doi: 10.1093/nar/gkq1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harris TW, Antoshechkin I, Bieri T, Blasiar D, Chan J, Chen WJW, De La Cruz N, Davis P, Duesbury M, Fang R, et al. WormBase: A comprehensive resource for nematode research. Nucleic Acids Res. 2010;38:D463–D467. doi: 10.1093/nar/gkp952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Elsworth B, Wasmuth J, Blaxter M. NEMBASE4: The nematode transcriptome resource. Int. J. Parasitol. 2011;41:881–894. doi: 10.1016/j.ijpara.2011.03.009. [DOI] [PubMed] [Google Scholar]
- 5.Blaxter ML, De Ley P, Garey JR, Liu LX, Scheldeman P, Vierstraete A, Vanfleteren JR, Mackey LY, Dorris M, Frisse LM, et al. A molecular evolutionary framework for the phylum Nematoda. Nature. 1998;392:71–75. doi: 10.1038/32160. [DOI] [PubMed] [Google Scholar]
- 6.Holterman M, van der Wurff A, van den Elsen S, van Megen H, Bongers T, Holovachov O, Bakker J, Helder J. Phylum-wide analysis of SSU rDNA reveals deep phylogenetic relationships among nematodes and accelerated evolution toward crown clades. Mol. Biol. Evol. 2006;23:1792–1800. doi: 10.1093/molbev/msl044. [DOI] [PubMed] [Google Scholar]
- 7.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 8.Parra G, Bradnam K, Korf I. CEGMA: A pipeline to accurately annotate core genes in eukaryotic genomes. Bioinformatics. 2007;23:1061–1067. doi: 10.1093/bioinformatics/btm071. [DOI] [PubMed] [Google Scholar]
- 9.Baird NA, Etter PD, Atwood TS, Currey MC, Shiver AL, Lewis ZA, Selker EU, Cresko WA, Johnson EA. Rapid SNP discovery and genetic mapping using sequenced RAD markers. PLoS ONE. 2008;3:e3376. doi: 10.1371/journal.pone.0003376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Andolfatto P, Davison D, Erezyilmaz D, Hu TT, Mast J, Sunayama-Morita T, Stern DL. Multiplexed shotgun genotyping for rapid and efficient genetic mapping. Genome Res. 2011;21:610–617. doi: 10.1101/gr.115402.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Davey JW, Hohenlohe PA, Etter PD, Boone JQ, Catchen JM, Blaxter ML. Genome-wide genetic marker discovery and genotyping using next-generation sequencing. Nat. Rev. Genet. 2011;12:499–510. doi: 10.1038/nrg3012. [DOI] [PubMed] [Google Scholar]