Abstract
The Death Domain (DD) superfamily, which is one of the largest classes of protein interaction modules, plays a pivotal role in apoptosis, inflammation, necrosis and immune cell signaling pathways. Because aberrant or inappropriate DD superfamily-mediated signaling events are associated with various human diseases, such as cancers, neurodegenerative diseases and immunological disorders, the studies in these fields are of great biological and clinical importance. To facilitate the understanding of the molecular mechanisms by which the DD superfamily is associated with biological and disease processes, we have developed the DD database (http://www.deathdomain.org), a manually curated database that aims to offer comprehensive information on protein–protein interactions (PPIs) of the DD superfamily. The DD database was created by manually curating 295 peer-reviewed studies that were published in the literature; the current version documents 175 PPI pairs among the 99 DD superfamily proteins. The DD database provides a detailed summary of the DD superfamily proteins and their PPI data. Users can find in-depth information that is specified in the literature on relevant analytical methods, experimental resources and domain structures. Our database provides a definitive and valuable tool that assists researchers in understanding the signaling network that is mediated by the DD superfamily.
INTRODUCTION
The Death Domain (DD) superfamily is one of the largest and most studied protein–protein interaction (PPI) modules and is comprised of four subfamilies: DD (1), Death Effector Domain (DED) (2), CAspase Recruitment Domain (CARD) (3) and PYrin Domain (PYD) (4). The functional and structural similarity are key feature for defining DD superfamily (1–4). The classification of the subfamily in the superfamily is mainly by sequence homology (3–7). Accumulating structural information, however, indicates that each subfamily has unique structural characteristics, such as a more flexible and exposed third helix in the DDs, the presence of an RxDL-motif in the DEDs, an interrupted first helix in the CARDs and relatively small third helix (or no well defined third helix) in the PYDs. All together, the sequence of amino acids and the structural characteristics defined the subfamily in the DD superfamily (5–7).
The DD superfamily plays a pivotal role in apoptosis, inflammation, necrosis and immune cell signaling pathways (5–9). Upon receiving signals to trigger apoptosis or inflammation signals, the assembly of caspase activating complexes occurs via the DD superfamily (10–12). The DD superfamily is also involved in recruiting downstream effectors for immune cell receptor signaling, intracellular pathogen sensing and DNA damage responses (13,14). Because DD superfamily-mediated signaling events are associated with various human diseases, such as cancers, neurodegenerative diseases and immunological disorders, the DD superfamily has emerged as a promising target for therapeutic intervention (15–17). A detail of the role and importance of DD superfamily in many cellular signaling pathways is beyond the scope of this article, and the reader is referred to recent review articles (5–9).
In the human genome, 37 proteins with DDs, 7 proteins with DEDs, 33 proteins with CARDs and 22 proteins with PYDs have been identified (5,6). Because specific DD superfamily-mediated PPIs are critical for determining downstream events, the investigation of PPIs among the DD superfamily proteins (i.e. proteins that contain the DD superfamily) will facilitate the understanding of DD superfamily-mediated molecular and cellular processes and their related diseases.
PPI modules, including the DD superfamily, have been intensively studied, because most proteins form complexes that achieve specific functions. Accumulating information on the PPIs in many cellular signaling pathways provides crucial insights into understanding the molecular mechanisms and their related disease processes. Because PPI interfaces have emerged as promising drug targets, there has been substantial progress in developing small molecular compounds that competitively interfere with PPIs (18).
Although there are several well-known global PPI network databases, such as DIP (19), IntAct (20), MINT (21) and STRING (22), these databases do not provide sufficient information to scientists who want to focus their research on specific protein families. Thus, it is desirable to construct extensively curated databases that can provide in-depth information on specific molecules or molecule families, and such databases will be more effective than the present global PPI network in stimulating the formulation of new knowledge, hypotheses or experiments (23).
Here, we present the DD database, a manually curated database that aims to provide comprehensive information on PPIs of the DD superfamily. This database has a user-friendly interface with many useful features, including a search engine, an interaction map and a function for cross-referencing useful external databases. Our DD database will provide a valuable tool to assist in understanding the molecular interaction and signaling network of the DD superfamily. The DD database can be accessed at http://www.deathdomain.org.
CONTENTS OF THE DATABASE
Our DD database contains 175 PPI pairs among the 99 DD superfamily proteins. The PubMed database has been used to collect information on the DD superfamily and its PPIs. After reviewing hundreds of articles, 295 peer-reviewed articles were manually selected to build the database. Users can access all the DD superfamily proteins, their PPI pairs and selected literature via the ‘Statistics’ page on the website (Supplementary Figure S1). An overview of the PPI information described in this article is presented in the form of a matrix (Table 1). Based on common research schemes of PPI studies (24), columns are labeled with the following three categorizations: validation for interaction, characterization and functional consequence (Supplementary Table S1). Validation for interaction is subdivided into in vitro and in vivo. These categorizations contain the answers for three basic but critical questions about PPIs. Validation and characterization will provide answers on how to identify and confirm PPIs and what are the biochemical properties of the PPIs, respectively. Functional consequence will present an answer to what are the biological meanings of the PPIs. Each category has a brief summary of experimental results or information on experimental resources.
Table 1.
A summary of the DD superfamily PPIs provided by the DD database
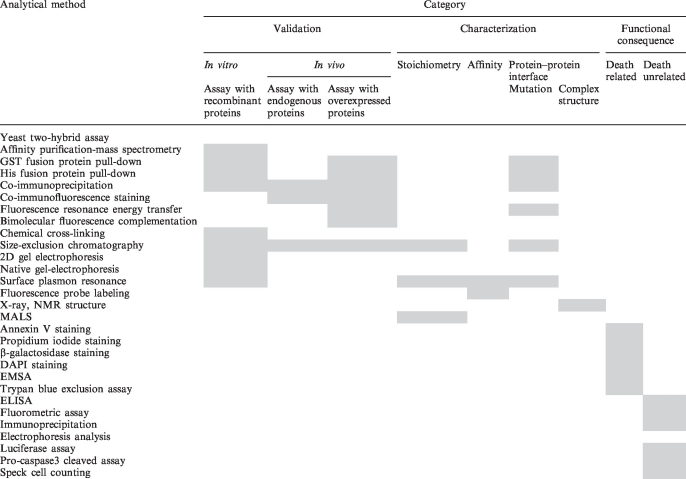 |
Analytical methods and their related categories are represented by gray boxes.
Validation refers to the type of experimental methods that are used to verify the DD superfamily PPIs. Validation for the PPIs is classified either as in vitro or in vivo. In vitro interaction refers to the methods that use assays with recombinant proteins, whereas in vivo interaction represents those processes that use assays with endogenous or overexpressed proteins. The DD database provides a detailed summary of experimental resources, including the gene constructs and expression systems that are used for recombinant protein preparation and cell/tissue types. Characterization refers to the data regarding the biochemical properties of PPIs, such as binding region mapping, stoichiometry or affinity among interacting proteins as specified in the articles. We curated the mapping data for binding regions with regard to the gene constructs that are used and corresponding amino acid regions. In the DD database, stoichiometry is described in quantitative terms and affinity is defined by dissociation constants. Because the DD superfamily proteins have apoptotic or non-apoptotic functions, we classified and curated their functional roles into two subcategories: death-related and death-unrelated.
The DD database provides information on the amino acid sequences and the domain boundaries of the DD superfamily proteins, which is retrieved from the UniProtKB/Swiss-Prot and UniProtKB/TrEMBL databases (25) and is available in FASTA, EMBL and GenBank formats (Supplementary Figure S2). External databases that introduce specific DD superfamily proteins and their PPIs are linked at the ‘External Database Link’ page on the website. Because the structural information of the DD superfamily is important for understanding PPIs at an atomic level, we also included any available 3D structure information of each DD superfamily. The details of procedures for structural determination are also contained in our database via the ‘3D Structure’ page. In addition, our database provides the information of natural mutations and related diseases via ‘Disease’ tab. Because the DD superfamily proteins are involved in many diseases through the certain types of mutation, this information might be particularly useful to understand mutation based diseases.
DATA COLLECTION AND CURATION
The PubMed database was used as the primary source for collecting information and constructing the DD database. After finding synonyms for each of the 99 DD superfamily proteins using UniProtKB (25) and Entrez Gene (26), we obtained a list of articles using each name of the proteins and its synonyms on a PubMed search, and we selected the articles that contained evidence for physical binding among the proteins denoted. We also manually screened information that was in other databases, such as DIP, IntAct, MINT, STRING and Entrez Gene. All of the 295 articles used for database construction are listed on our database website (Supplementary Figure S1).
We curated the data that were based on analytical methods, experimental results, resources (e.g. genes or proteins, primary cells, tissues and cell lines) and nomenclature. The curation of the analytical methods and experimental results was partly described in the previous section ‘CONTENTS OF DATABASE’. We manually curated the information on the gene constructs that were denoted in the literature with respect to species and amino acid region. With regard to the nomenclature that describes the proteins, we adopted common names that are widely used in the literature. If information on the gene constructs was not found in the literature, but relevant references were provided, we gathered information from the referenced articles. However, if no appropriate data were available, the entry would be designated as ‘Not specified’ in our database. Because the nomenclatures describing primary cells or tissues are mostly unified, we faithfully cited primary cell or tissue names that were designated in the literature.
WEB INTERFACES
The web interfaces to the DD database were developed using XHTML 1.1, CSS, JavaScript and Ruby on Rails 2.3.8 and runs on a Phusion Passenger application server (version 2.2.15) with an Apache HTTP server (version 2.2.14) hosted on an Ubuntu Linux server (version 10.04.3 LTS). All of the data for the DD database were stored on a MySQL server (version 5.1.41). The web-based network visualization tool of the DD database was developed using Adobe Flex 4 SDK and Flare. Jmol was also used for the visualization of 3D structures of proteins (27).
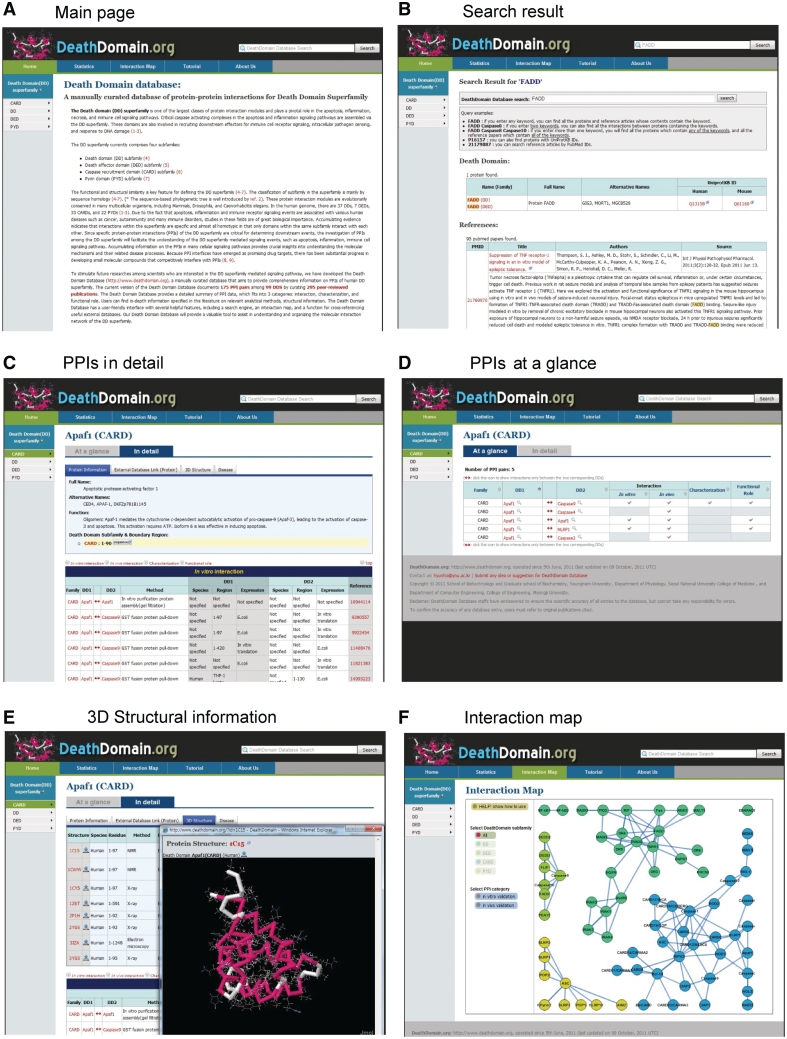
The DD database contains several useful features, such as a search engine, an interaction map and a tool that cross-references other external resources. Navigation of the DD database is illustrated in Figure 1, and the website also provides instruction in the ‘Tutorial’ section. Users can browse the database by clicking the name of a subfamily of a DD superfamily (CARD, DD, DED and PYD) or the name of an individual the DD superfamily protein. If users choose to begin their search based on a specific subfamily of the DD superfamily, they will obtain all the information that is related to a subfamily containing proteins, including lists of PPI pairs of the subfamily and the availability of data for the validation of the interaction, characterization and functional roles of each PPIs in the ‘At a glance’ section on the website (Supplementary Figure S3). Users also can access all of the detailed PPI information of a certain DD superfamily by clicking the ‘In detail’ section on the website (Supplementary Figure S3). If users choose to browse based on a specific name for the DD superfamily proteins, they will see the information on the brief introduction of the proteins, amino acid sequences, domain boundaries and PPIs on the front page (Figure 1C and D). Users also can click on the arrow between two interacting DD members to get information on the specific interactions. UniProtKB IDs and availability of 3D structures of a specific DD superfamily also can be accessed by clicking ‘External Database Link’ and ‘3D structure’, respectively (Figure 1E).
Figure 1.
Overview of the DD database web interfaces, with a reconstituted illustration. (A) Main page (B) Search result (C) PPI in detail (D) PPIs at a glance (E) 3D structure information (F) Interaction map.
The DD database provides a full-text search tool for searching target proteins, PPIs and relevant literatures (Figure 1B). When users enter a query (i.e. protein names or UniProtKB IDs) into the search form, the system presents corresponding information on all of the proteins and PPI pairs. In addition, the system can provide a list of indexed entries that contain the search word in the title, author, affiliation or abstract. The DD database also provides graphical displays for the complicated network of DD superfamily-mediated PPIs for better visualization. The proteins are represented as round-shaped nodes that are labeled with their names, and PPIs are illustrated as edges connecting the protein nodes (Figure 1F). By clicking the nodes, all of the PPI pairs that are generated by a specific protein become highlighted. Detailed information on a PPI can be obtained by double-clicking the representative node. Users can also drag the nodes for better visualization. A control panel on the left side of the toolbar allows users to choose each of the DD subfamilies and PPI categories.
To complement the DD database and to provide additional information on each gene that is contained in the DD database, we provide hyperlinks to other useful databases, including UniProtKB, DIP, IntAct, MINT, STRING and KEGG. Thus, the DD database contributes as a central hub for the collection of structural, functional and signaling information on the DD superfamily and its PPIs in detail.
DISCUSSION AND FUTURE DIRECTIONS
Studies on PPI modules and their PPIs in many cellular signaling pathways are critical for understanding the molecular mechanisms and their related disease processes. Because publications that are related to the DD superfamily-mediated signaling pathways have increased remarkably in recent years, additional effort to organize the comprehensive information of the DD superfamily and its PPIs is still needed. In addition, because global PPI databases do not provide sufficient information to scientists who want to focus their research on specific protein families, it is desirable to construct specific databases that contain detailed information on relevant analytical methods, experimental resources and summarized results.
Our DD database provides in-depth information on the DD superfamily and its PPIs, which assists in developing an understanding of the molecular and signaling network that is mediated by the DD superfamily. Our database provides documentation on the experimentally validated PPIs among the DD superfamily proteins. Manual curation is currently the best method for constructing reliable biological databases and is also able to include all of the detailed information, such as the species of the genes, the domain boundaries of the proteins, the relevant experimental methods and the structural information. The superiority and accuracy of the manual curation become even clearer when our database was compared with other PPI databases (Supplementary Table S2). Therefore, the aim of this database is to provide the scientific community with a comprehensive and integrated tool for efficiently, conveniently and accurately extracting information about PPIs of the DD superfamily
We are also planning to further extend the contents of our database, to add to the DD superfamily. Because several more cell death-related domains, including the CIDE, BH3 and BIR domains, have been identified and characterized, we plan to include these domains in the content of our DD database. Additionally, we are developing computational methods for discovering unidentified or possible PPI pairs using network analysis tools. Finally, we believe that our database constitutes a step toward optimizing and generalizing the use of the DD superfamily PPI search tools to be accessible beyond the specialized research community.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR online: Supplementary Figures 1–3 and Supplementary Tables 1 and 2.
FUNDING
Funding for open access charge: Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science and Technology (2011-0003406 and 2011-0025697 to H.H.P., 2008-05943 to I.S. and 2011-0022437 to D.K.).
Conflict of interest statement. None declared.
REFERENCES
- 1.Tartaglia LA, Ayres TM, Wong GHW, Goeddel DV. A Novel Domain within the 55 kd TNF Receptor Signals Cell Death. Cell. 1993;74:845–853. doi: 10.1016/0092-8674(93)90464-2. [DOI] [PubMed] [Google Scholar]
- 2.Chinnalyan AM, O'Rourke K, Tewari M, Dixit VM. FADD, a novel death domain-containing protein, interacts with the death domain of FAS and initiates apoptosis. Cell. 1995;81:505–512. doi: 10.1016/0092-8674(95)90071-3. [DOI] [PubMed] [Google Scholar]
- 3.Hofmann K, Bucher P, Tschopp J. The CARD domain: a new apoptotic signaling motif. Trends Biochem. Sci. 1997;22:155–156. doi: 10.1016/s0968-0004(97)01043-8. [DOI] [PubMed] [Google Scholar]
- 4.Martinon F, Hofmann K, Tschopp J. The pyrin domain: a possible member of the death domain-fold family implicated in apoptosis and inflammation. Curr. Biol. 2001;11:R118–R120. doi: 10.1016/s0960-9822(01)00056-2. [DOI] [PubMed] [Google Scholar]
- 5.Park HH, Lo YC, Lin SC, Wang L, Yang JK, Wu H. The Death Domain superfamily in intracellular signaling of apoptosis and inflammation. Ann. Rev. Immunol. 2007;25:561–586. doi: 10.1146/annurev.immunol.25.022106.141656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reed JC, Doctor KS, Godzik A. The domains of apoptosis: a genomics perspective. Sci. STKE. 2004;2004:re9. doi: 10.1126/stke.2392004re9. [DOI] [PubMed] [Google Scholar]
- 7.Kersse K, Versputen J, Berghe TV, Vandenabeele P. The death-fold superfamily of homotypic interaction motifs. Trends Biochem. Sci. 2011;36:541–552. doi: 10.1016/j.tibs.2011.06.006. [DOI] [PubMed] [Google Scholar]
- 8.Bertin J, DiStefano PS. The PYRIN domain: a novel motif found in apoptosis and inflammation proteins. Cell Death Differ. 2000;7:1273–1274. doi: 10.1038/sj.cdd.4400774. [DOI] [PubMed] [Google Scholar]
- 9.Martinon F, Hofmann K, Tschopp J. The pyrin domain: a possible member of the death domain-fold family implicated in apoptosis and inflammation. Curr. Biol. 2001;11:R118–R120. doi: 10.1016/s0960-9822(01)00056-2. [DOI] [PubMed] [Google Scholar]
- 10.Bratton SB, Walker G, Srinivasula SM, Sun XM, Butterworth M, Alnemri ES, Cohen GM. Recruitment, activation and retention of caspases-9 and -3 by Apaf-1 apoptosome and associated XIAP complexes. EMBO J. 2001;20:998–1009. doi: 10.1093/emboj/20.5.998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chou JJ, Matsuo H, Duan H, Wagner G. Solution structure of the RAIDD CARD and model for CARD/CARD interaction in caspase-2 and caspase-9 recruitment. Cell. 1998;94:171–180. doi: 10.1016/s0092-8674(00)81417-8. [DOI] [PubMed] [Google Scholar]
- 12.Martinon F, Burns K, Tschopp J. The inflammasome: a molecular platform triggering activation of inflammatory caspases and processing of proIL-beta. Mol. Cell. 2002;10:417–426. doi: 10.1016/s1097-2765(02)00599-3. [DOI] [PubMed] [Google Scholar]
- 13.Lin SC, Lo YC, Wu H. Helical assembly in the MyD88-IRAK4-IRAK2 complex in TLR/IL-1R signalling. Nature. 2010;465:885–890. doi: 10.1038/nature09121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Muzio M, Ni J, Feng P, Dixit VM. IRAK (Pelle) family member IRAK-2 and MyD88 as proximal mediators of IL-1 signaling. Science. 1997;278:1612–1615. doi: 10.1126/science.278.5343.1612. [DOI] [PubMed] [Google Scholar]
- 15.Rieux-Laucat F, Fischer A, Deist FL. Cell-death signaling and human disease. Curr. Opin. Immunol. 2003;15:325–331. doi: 10.1016/s0952-7915(03)00042-6. [DOI] [PubMed] [Google Scholar]
- 16.Smahi A, Courtois G, Rabia SH, Doffinger R, Bodemer C, Munnich A, Casanova JL, Israel A. The NF-kappaB signalling pathway in human diseases: from incontinentia pigmenti to ectodermal dysplasias and immune-deficiency syndromes. Hum. Mol. Genet. 2002;11:2371–2375. doi: 10.1093/hmg/11.20.2371. [DOI] [PubMed] [Google Scholar]
- 17.Coban C, Ishii KJ, Akira S. Immune Interventions of Human Diseases through Toll-Like Receptors. Adv. Exp. Med. Biol. 2009;655:63–80. doi: 10.1007/978-1-4419-1132-2_7. [DOI] [PubMed] [Google Scholar]
- 18.Wells JA, McClendon CL. Reaching for high-hanging fruit in drug discovery at protein-protein interfaces. Nature. 2007;450:1001–1009. doi: 10.1038/nature06526. [DOI] [PubMed] [Google Scholar]
- 19.Salwinski L, Miller CL, Smith AJ, Pettit FK, Bowie JU, Eisenberg D. The Database of Interacting Proteins: 2004 update. Nucleic Acids Res. 2000;32:D449–D451. doi: 10.1093/nar/gkh086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aranda B, Achuthan P, Alam-Faruque Y, Armean I, Bridge A, Derow C, Feuermann M, Ghanbarian AT, Kerrien S, Khadake J, et al. The IntAct molecular interaction database in 2010. Nucleic Acids Res. 2010;38:D525–D531. doi: 10.1093/nar/gkp878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ceol A, Chatr Aryamontri A, Licata L, Peluso D, Briqanti L, Perfetto L, Castagnoli L, Cesareni G. MINT, the molecular interaction database: 2009 update. Nucleic Acids Res. 2010;38:D532–D539. doi: 10.1093/nar/gkp983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Szklarczyk D, Franceschini A, Kuhn M, Simonovic M, Roth A, Minquez P, Doerks T, Stark M, Muller J, Bork P, et al. STRING: a database of predicted functional associations between proteins. Nucleic Acids Res. 2011;39:D561–D568. doi: 10.1093/nar/gkq973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shin YC, Shin SY, So I, Kwon D, Jeon JH. TRIP Database: a manually curated database of protein-protein interactions for mammalian TRP channels. Nucleic Acids Res. 2011;39:D356–D361. doi: 10.1093/nar/gkq814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xenarios I, Eisenberg D. Protein interaction databases. Curr. Opin. Biotechnol. 2001;12:334–339. doi: 10.1016/s0958-1669(00)00224-x. [DOI] [PubMed] [Google Scholar]
- 25.The UniProt Consortium. The Universal Protein Resource (UniProt) in 2010. Nucleic Acids Res. 2010;38:D142–D148. doi: 10.1093/nar/gkp846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maglott D, Ostell J, Pruitt KD, Tatusova T. Entrez Gene: gene-centered information at NCBI. Nucleic Acids Res. 2011;39:D52–D57. doi: 10.1093/nar/gkq1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Herraez A. Biomolecules in the computer: Jmol to the rescue. Biochem. Mol. Biol. Educ. 2006;34:255–261. doi: 10.1002/bmb.2006.494034042644. [DOI] [PubMed] [Google Scholar]