Abstract
The TDR Targets Database (http://tdrtargets.org) has been designed and developed as an online resource to facilitate the rapid identification and prioritization of molecular targets for drug development, focusing on pathogens responsible for neglected human diseases. The database integrates pathogen specific genomic information with functional data (e.g. expression, phylogeny, essentiality) for genes collected from various sources, including literature curation. This information can be browsed and queried using an extensive web interface with functionalities for combining, saving, exporting and sharing the query results. Target genes can be ranked and prioritized using numerical weights assigned to the criteria used for querying. In this report we describe recent updates to the TDR Targets database, including the addition of new genomes (specifically helminths), and integration of chemical structure, property and bioactivity information for biological ligands, drugs and inhibitors and cheminformatic tools for querying and visualizing these chemical data. These changes greatly facilitate exploration of linkages (both known and predicted) between genes and small molecules, yielding insight into whether particular proteins may be druggable, effectively allowing the navigation of chemical space in a genomics context.
BACKGROUND
The open access, web accessible TDR Targets database (http://tdrtargets.org) (1), allows users to interrogate pathogen specific genomic-scale information and to identify and prioritize high value targets based on whether or not they fulfill a set of user defined criteria. The name of the database includes the initialism ‘TDR’ for Tropical Disease Research, a special program within the World Health Organization (see Acknowledgements). The focus of the TDR Targets database is on high priority tropical disease pathogens (currently the top ten pathogens in the portfolio of this special program), and several other phylogenetically relevant pathogens (the Wolbachia endosymbiont of Brugia malayi and the apicomplexan parasite Toxoplasma gondii, for example). The database integrates information on gene products from primary genome databases (2–6), and gathers, from various resources and published studies, organism-specific functional information such as information on orthologues (7), 3D structures (8) and/or structural models [modeling of pathogen proteins were obtained for this work, and are now available from Modbase (9)], enzyme/metabolic pathway classification, expression and essentiality (1). These datasets are further supplemented with information curated from the literature on chemical and/or genetic validation status of targets, precedence for druggability and assayability. Ultimately, the genomic scale datasets compiled in TDR Targets should aid the tropical infectious disease community in driving drug discovery efforts. The importance of this is obvious, given the urgent need for new drugs, the rapid emergence of resistance and toxicity issues associated with existing drugs, and considering that the drug discovery pipeline for tropical infectious diseases is rather thin due to the chronic underfunding of the field and a lack of commercial interest from big pharmaceutical companies.
Combining genomics data with chemical data is essential for the success of discovery efforts. The availability of large compound datasets is particularly important in tropical infectious disease research. Until recently, however, access to large-scale pharmacological and medicinal chemistry datasets was limited (mostly due to the proprietary nature of the data), or prohibitively expensive. However, this landscape started to change dramatically in the past 5 years with the advent of a number of open access chemical resources such as PubChem, ChEMBL and others (10), and by the release of key high-throughput screening datasets by both academic and pharmaceutical companies (11,12) (Novartis-GNF Malaria Box, K Gagaring, R Borboa, C Francek, Z Chen, J Buenviaje, D Plouffe, E Winzeler, A Brinker, T Diagana, J Taylor, R Glynne, A Chatterjee, K Kuhen. Genomics Institute of the Novartis Research Foundation (GNF) USA, and Novartis Institute for Tropical Disease, Singapore).
In order to complement the genomic datasets and target-focused functionalities available in TDR Targets (1), we integrated in the database a number of chemical datasets enriched in drugs and drug-like molecules collected from various sources, and developed cheminformatics components that drive a data loading pipeline and parts of the web application, allowing users to mine these data. In this report, we describe these new functionalities and data, and how the integration of chemical and genomics data can be used to formulate relevant queries to identify either new chemical leads for a target, or candidate new targets for orphan bioactive molecules.
Several existing databases allow searching for target and/or drug information, but each of these has been designed for a different purpose. Some are focused predominantly on describing chemical entities, while others are focused on specific aspects of proteins that make them possible targets, such as position in a metabolic pathway or structural features. Examples of databases focused on chemical entities include ChemBank (13), which is focused on small molecule activities, obtained from a wide variety of screenings; ChEBI (Chemical Entities of Biological Interest) (14), which contains chemical information and properties of small molecules; and Drugbank (15) which holds chemical data on FDA approved drugs, nutraceutical and experimental drugs, together with information about their related proteins (targets, enzymes, etc.). Others like BindingDB (16) catalogue experimentally validated protein–ligand interactions relevant to drug discovery and provide detailed information on binding affinities and inhibitory kinetics. The SuperDrug Database (17) allows users to search and compare structures of approved drugs, while the Antimicrobial peptide database (18) also allows users to browse or search for peptides that target infectious agents or cancer cells based on their activity and content. There are several databases focused primarily on protein targets—most of these focus on a single prioritization theme, or a single organism. For example, the Genomic Target Database (19) contains lists of drug targets for four human bacterial pathogens, with specific lists of targets grouped by metabolic pathway or membrane localization. The Potential Drug Target Database (PDTD) (20), allows users to browse solved structures of potential drug targets and to identify potential targets of a given small molecule through docking into many solved structures. The Therapeutic Targets Database (21) contains a large volume of data on known and predicted targets, allowing users to download target validation data for individual proteins, to view Quantitative Structure-Activity Relationship (QSAR) data for individual proteins, or to search targets by linked compounds. Finally, there are a number of cheminformatics resources such as HEOS (22) that facilitate the remote collaboration between groups involved in a drug discovery process. However, very few of these databases offer the same breadth of data-types relevant to drug target characterization as TDR Targets. ChEMBL and PubChem are two examples of chemical databases that provide additional links to protein targets. Nonetheless, TDR Targets stands alone in its ability to allow users to frame weighted, complex queries to rank and interrogate many targets at once from different species, and to cross-relate chemical data with target data in meaningful ways, as described in this report. In this respect we believe TDR Targets fills an important niche. To our knowledge, the TDR Targets database represents a unique resource providing comprehensive compilation of relevant data for drug target prioritization on multiple pathogens, all in one place.
NEW FUNCTIONALITY: INTEGRATION OF CHEMICAL DATA IN TDR TARGETS
In order to collect chemical information that would be useful to search for new bioactive compounds against tropical diseases, we selected data sources enriched in drugs and drug-like molecules. Chemical compounds listed in our database come from: the ChEMBL database (http://www.ebi.ac.uk/chembldb), that contains information on small molecules together with bioactivity information curated from the literature, and information on associated protein targets (23) (443 602 compounds); PubChem (24) (278 070 compounds); the DrugBank database, which contains information on FDA approved drugs (15) (4421 compounds). The Tres Cantos Antimalarial TCAMS dataset (GSK) (13 469 compounds) (11), the Novartis-GNF Malaria Box database (5388 compounds), and the St. Jude Children's Research Hospital Malaria dataset (305 815 compounds)(12), three datasets that contain molecules tested in high-throughput screening assays against Plasmodium falciparum, were obtained from ChEMBL-NTD (http://www.ebi.ac.uk/chemblntd), a repository of screening data of molecules directed against neglected diseases.
A total of 825 814 unique drug-like compounds are present in the combined dataset obtained from the above indicated sources. These were integrated into the TDR Targets database along with data on: (i) basic information such as chemical name, InChi and InChiKey identifiers; (ii) chemical properties (molecular weight, logP, hydrogen bond donors, hydrogen bond acceptors, flexible bonds, Lipinski rule of 5 compliance); (iii) chemical structure; (iv) bioactivities (IC50, Ki, MIC, Activity, EC50, ED50 and percent growth inhibition, among others) and (v) association with genes (curated, predicted or any). Table 1 provides a summary of searchable fields for chemical data. Users can access this information either by searching for target genes, and then looking up the chemicals linked to these genes (scenario A in Figure 1) or by searching the chemical space itself (scenario B in Figure 1).
Table 1.
Searchable chemical information in TDR Targets
| Data types | Format / Example / Observations | |
|---|---|---|
| Structure | 2D structure | SDF / MOL |
| Identifiers | InChI | 1S/C7H11NO7P2/c9-7(16(10,11)12,17(13,14)15)4-6-2-1-3-8-5-6/h1-3,5,9H[…] |
| InChI Key | IIDJRNMFWXDHID-UHFFFAOYSA-N | |
| ChEMBL | 183772 | |
| DrugBank | DB00884 | |
| PubChem CID | 5245 | |
| Textual information | Name/ Synonyms | Risedronate / 1-hydroxy-1-phosphono-2-pyridin-3-ylethyl) phosphonic acid |
| Data Source | ChEMBL, PubChem, DrugBank | |
| Chemical properties | MW | 282.104 |
| Formula | C7H10NO7P2 | |
| LogP | −3.23 | |
| No. of H donors | 4 | |
| No. of H acceptors | 8 | |
| No. of flexible bonds | 4 | |
| Activity | Assay | e.g. Inhibitory activity against Leishmania major Farnesyl diphosphate synthase |
| Readout | IC50, EC50, Ki, MIC, % growth inhibition, etc. (depends on particular assay) | |
| Target association | Direct | Manual curation (experimental evidence, target directly assayed) |
| Transitive | Experimental evidence available for an ortholog/homolog |
Searchable information fields for small molecules integrated in TDR Targets are shown in the Table. The 2D structure of a molecule is used for similarity and substructure searches, and in all searches started from the JME molecule editor. All other information fields are searchable as textual or numeric information using standard forms (see Figure 3 for some examples). Target associations are used internally to limit search results to show only those compounds that are associated with a target (see examples in Figure 2), and to display links to targets within pages. Note: the InChI string in the table has been truncated for presentation purposes.
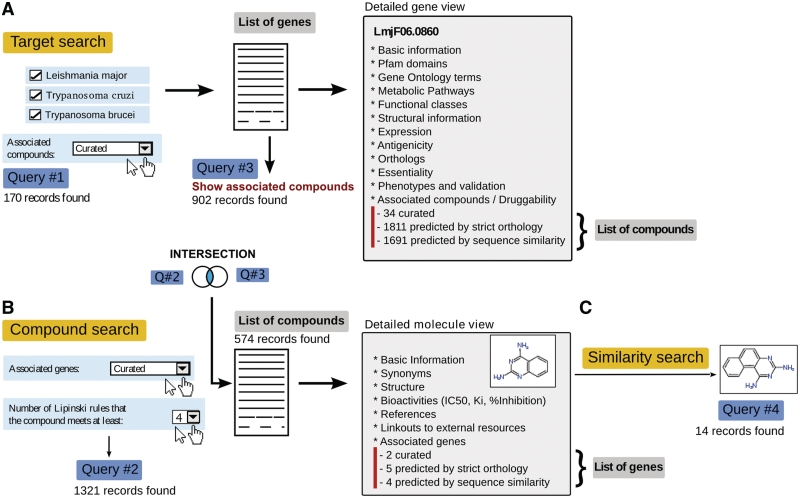
Figure 1.
Schematic view of chemogenomic searches and navigation supported in TDR Targets. (A) Targets search. Query #1 retrieves 170 genes from Leishmania major, Trypanosoma cruzi and Trypanosoma brucei that were associated to compounds by manual curation. Clicking on gene LmjF06.0860 (dihydrofolate reductase-thymidylate synthase from L. major) shows the corresponding gene page, and allows users to inspect the associated compounds. In TDR Targets a target resultset can be used to generate the corresponding compound resultset by clicking on the ‘Show associated compounds' link (and vice versa for compound resultsets). Query #3 was generated in this way, and produces a list of 902 compounds associated to trypanosomatid genes by manual curation. (B) Compound search (textual). Query #2 was performed from the compounds search page, retrieving 1321 compounds that meet all 4 of Lipinski's rules, and that were associated to genes by manual curation. The combination of queries #2 and #3 (INTERSECTION) can be calculated at the history page, returning 574 compounds that meet all specified criteria. (C) Compound similarity search. In Query #3, a 2,4-diaminoquinazoline was found associated to 2 trypanosomatid genes by manual curation. In order to find additional related compounds with potential activity against this target, a similarity search can be performed (Query #4), retrieving another 14 compounds at Tanimoto similarity ≥0.8 (chemical analogs).
In a target search, it is now possible to query for targets with associated compounds and to further filter this search based on evidence for association (i.e. curated or predicted, or any). After running any target search, a list of targets is shown with links to all compounds associated with all the targets present in the list (Figure 1).
From the compound search page (http://tdrtargets.org/drugs) users can query chemical information using either text-based searches or structure-based searches (Figures 1 and 2) to retrieve a list of matching compounds. Users can then choose to either view details of individual compounds in the list (by clicking on the relevant compound) or retrieve all genes associated with the compounds listed using the ‘show all/curated/predicted genes’ link. In the first case, a new page is shown, with a detailed view for the compound, including basic chemical information, synonyms, structure, association with targets (and kind of association), activities described for the compound, external resources, and bibliographic references. In the second case, a new query is run to search for all/curated/predicted pathogen genes associated with all the compounds present in the list. The retrieved genes are shown in similar fashion to the result for a typical target search (Figure 1). All the history functionalities, such as combining query results using Boolean AND, OR and NOT operators as well as saving, exporting and publishing query results, that were previously available for handling target searches are also available for handling chemical data searches.
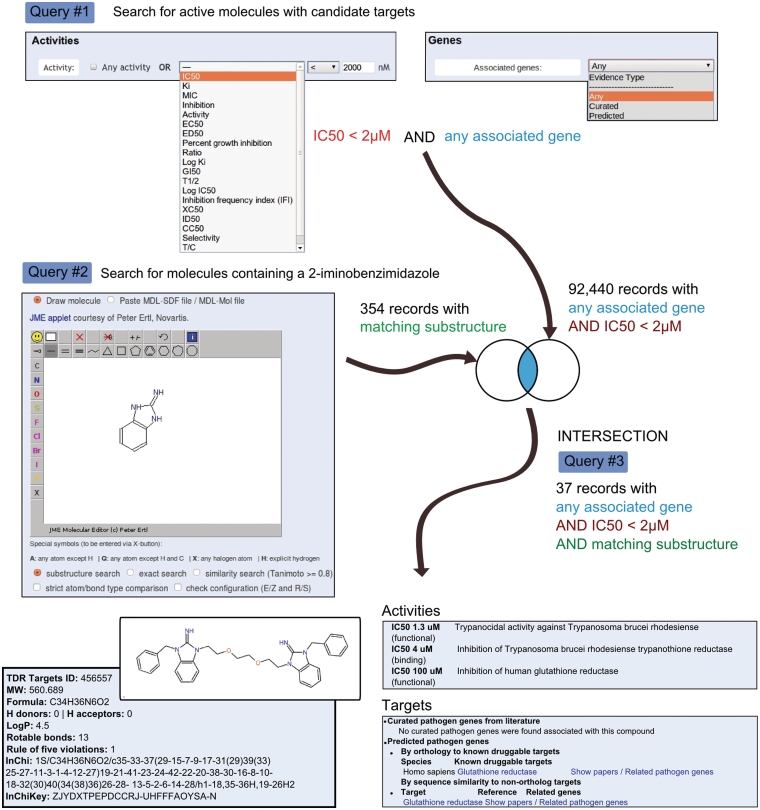
Figure 2.
A query strategy designed to find active compounds from a defined chemical class. TDR Targets allows searches of the activity of compounds. Query #1 uses this functionality to retrieve reasonably active chemical leads that have been associated to targets. TDR Targets also allows users to perform substructure searches. Query #2 implements such a search, retrieving compounds containing the drawn structure as part of the molecule. The intersection of these two queries finds 37 active compounds from this class (2-iminobenzimidazoles). One example molecule from this list is shown at the bottom, with a few selected panels of information from the corresponding compound page.
CONNECTING KNOWN DRUG TARGETS TO PATHOGEN GENOMES
Drug repositioning or repurposing, i.e. finding a new indication for existing drugs or drug-like molecules, can greatly speed up the traditional drug discovery process, which typically takes more than a decade to complete (25). An open access chemogenomics resource such as TDR Targets can now provide neglected disease researchers and pharma companies with a basic tool for knowledge-based drug repositioning (in a general sense). A key element of such a chemogenomics approach is the linking of target genes to suitable chemical inhibitors, and the leveraging of other relationships available in the database (e.g. sequence similarity between targets; chemical similarity between compounds).
As mentioned above, TDR Targets now contains a chemical database of bioactive compounds and their targets from primary data sources such as ChEMBL, DrugBank and PubChem, in addition to data collected from high-throughput screenings (The Tres Cantos Antimalarial TCAMS dataset, the Novartis-GNF Malaria Box database, the St. Jude Children's Research Hospital Malaria dataset), and in-house curatorial efforts by the TDR Targets team. Many of the compounds included from the above sources into TDR Targets have known target genes, mostly from non-pathogens, and this information can be used to link the compounds to pathogen target genes using in silico approaches such as orthologue mapping and protein domain conservation between the known targets and novel targets. In this way, users of the TDR Targets resource can find potential pathogen drug targets, linked to a set of chemical compounds with measured activity against a related protein. From here, chemical scaffolds of the proposed compounds can be used as starting points to identify new chemical entities as potential drugs for the novel target. A schematic view of this approach is presented in Figure 1.
The strategy of mapping gene functional data and chemical bioactivity data across orthologues is a key mechanism by which TDR Targets establishes links between chemicals and target genes. Orthologues are a set of genes from two or more species that originated by vertical descent from a single gene in the last common ancestor. Orthologues are often functionally and structurally similar. Thus, they may be modulated by identical or similar molecules, making orthology assessment a powerful tool to connect a known druggable target with a potential novel target [see (26) for an example of this strategy]. However, non-orthologous genes can share homologous druggable domains. Methods to predict orthology such as COG (27) and OrthoMCL (7) are based on a reciprocal best BLAST hit step (i.e. the first sequence finds the second sequence as its best hit in the second species, and vice versa). Protein coding genes containing multiple domains with one of them being ‘druggable’ could pass undetected because of the reciprocal best hit requirement of orthology-based methods (even if the other domains are conserved, but rearranged in the protein sequence). Thus, we implemented a second mapping strategy in TDR Targets using BLAST, in which a sequence similarity search is used to identify drug targets with high similarity to pathogen genes (although not necessarily reciprocal), with the requirement that the similarity span should almost completely cover the ‘druggable’ target (≥80% coverage, E-value <10−10). With this simple approach we are able to connect members of a protein family, which may be grouped in different ortholog clusters, although they are structurally similar. Using this strategy in the TDR Targets database, 3575 known druggable targets (primary drug-target association is based on manual curation) were assigned to 1509 OrthoMCL clusters of orthologous genes. About 50% of these groups (798) contain 4529 genes from tropical disease pathogens (and are therefore candidate druggable genes). The additional sequence similarity step performed with BLAST allowed us to identify 2087 pathogen genes that were not detected by strict orthology. A number of example cases are depicted in Figure 3. In TDR Targets, these similarity links between targets provide additional navigation routes in the database, facilitating the assessment of available evidence (e.g. activity of compounds) for a related group of homologues.
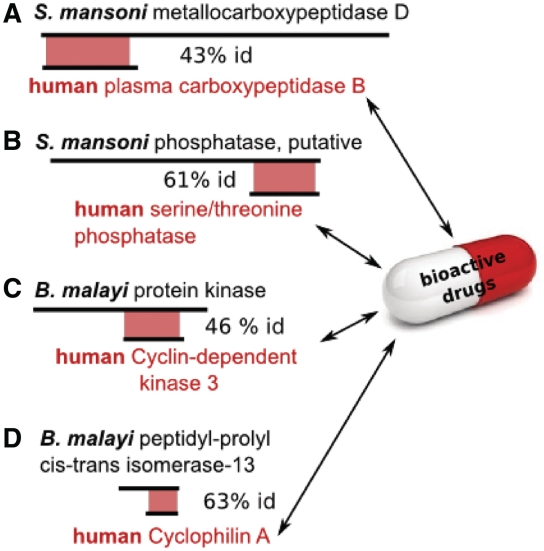
Figure 3.
Links between non-homologous genes in TDR Targets. Many druggable target sequences can be completely aligned to pathogen genes for which no druggable orthologs can be detected using stricter orthology methods (generally due to large differences in protein length between homologs). The figure depicts a number of schematic views of alignments between druggable targets (names shown in red) and helminth genes (% id = percentage identity). Genes represented from top to bottom (OrthoMCL Ortholog Cluster Identifier in brackets, IC50 of most active compound in square brackets) are: (A) S. mansoni Smp_159890 (OG4_13640), H. sapiens P15169 (OG4_27945) [2nM, PubChem CID 194328]; (B) Smp_155200 (OG4_12097), P62136 (OG4_10262) [0.1 nM, PubChem CID 445434]; (C) Bm1_17240 (OG4_12720), Q00526 (OG4_10184) [5 µM, PubChem CID 4369491]; (D) Bm1_52100 (OG4_12799), P62937 (OG4_10089) [2 nM, PubChem CID 9855081].
QUERYING THE AVAILABLE CHEMICAL INFORMATION IN TDR TARGETS
The new chemical data available in TDR Targets can be queried in a number of different ways. Every compound entry in TDR Targets is associated with a number of searchable parameters such as basic information (name, synonyms, InChi and InChi Key identifiers, data source), chemical properties (formula and atomic composition, molecular weight, solubility [logP], Lipinski rule of 5 compliance), bioactivity (if applicable), target genes (if applicable), and chemical structure (Table 1). At the top of the compound search page (Figure 2), the user can perform queries based on basic information, chemical properties, number and type of atoms, activities, association with genes and type of association (curated, predicted or any), and information source (Table 1). The InChi and InChiKey identifiers are unique representations of a molecule that can be used to look for a specific compound of interest. Alternatively, chemical properties like the molecular weight, logP, etc., are useful to retrieve a collection of compounds that meet user-defined criteria. Users can search for desirable compounds using a combination of search parameters. For example, one can search for molecules that have a molecular weight below 500, and ‘any’ described activity, or for compounds that are associated with genes by manual curation of literature, and meet all four Lipinski rules. An illustration of this type of chemical data search using various parameters is shown in Figure 3.
Searching for specific compounds using names or synonyms is possible in TDR Targets, however, these types of searches often do not produce the expected results, in many cases due to the different names and synonyms available for a molecule, as well as possible alternative spellings (e.g. metrifonate versus metriphonate). A more useful approach would be to use the 2D chemical structure of the compound to run an ‘exact match’ search to retrieve only the compound being searched for. In TDR Targets these type of searches can be initiated by drawing a molecule in the ‘Structure-based searches’ section of the compounds search page (http://tdrtargets.org/drugs). Users can draw molecule structures using the JME Java applet integrated in TDR Targets (JME molecule editor courtesy of Peter Ertl, Novartis http://www.molinspiration.com/jme/index.html), and run ‘exact’, ‘similarity’, or ‘substructure’ searches. This latter type of search will find any compound that contains the drawn structure as a part of it, and is usually a good way to find molecules that belong to a given chemical class (e.g. 2-iminobenzimidazoles, see Figure 3 for an example), for example to analyze and compare their bioactivities. Another potential use of this tool is to exclude certain compounds from a search, for example those containing undesirable functional groups (e.g. reactive/toxic in vivo, metabolically unstable or known to cause problems in screening for a particular assay). In such a case, the users could first query the database using an initial set of criteria (e.g. search for all compounds that have a measured IC50 below 2 µM). Then, a second query would retrieve all the compounds that contain an undesirable substructure. Finally, the second query can be subtracted from the first at the query history page, therefore obtaining a list of compounds that meet the first criteria (IC50 below 2 µM) but lack the undesirable substructure. Another way of finding potentially active molecules starting from a known bioactive compound is by similarity searching. To measure the similarity between the molecules, TDR Targets implements the Tanimoto (Jaccard) association coefficient which is a commonly used metric for chemical similarity of small molecules (28,29). Molecules that have a Tanimoto index equal to or greater than 0.8 to the query molecule are retrieved. If the molecule is too large to be easily drawn, but the structure is available as a molfile (in SDF/MOL format), the same searches can be initiated by pasting the content of the molecule's molfile (text) into the corresponding input textbox.
Altogether, these implemented query modes allow users to find either individual compounds or groups of compounds for later inspection of the information associated with them, including targets. Links to other resources such as ChEMBL, PubChem and ChemSpider (30) allow users to gather additional information on compounds.
USE CASES: SOME QUESTIONS THAT CAN BE ANSWERED USING THE NEW FUNCTIONALITIES
The new chemical functionalities developed and the recent integration of chemical data allow users to answer questions about compounds directly (e.g. in the compound search page), and also formulate questions about pathogen targets (from the targets search page, where the chemical information can be used as an additional criteria to restrict genes searches). Relevant questions that are now possible in TDR Targets include the following: find compounds with activity against Leishmania that are also associated with genes based on manual curation of the literature. This question can be answered by performing a single query in the compounds search page, selecting those compounds with assay descriptions matching ‘leishmania' (‘Description' box in the ‘Activities’ section), and choosing ‘Curated' in the ‘Gene Associations' section. This query results in a list of 63 compounds, that includes drugs such as pamidronate, risedronate, artemisinine, oryzalin and suramin.
Another example helps to illustrate how the integration of genomics and chemical data can be used to select candidate druggable targets. In this case, the following question can be easily translated into a search strategy in TDR Targets: which P. falciparum genes have evidence of essentiality in any species (through orthology), and have associated compounds by manual curation of literature? The corresponding search strategy would start by querying the database from the target search page, first choosing the species (Plasmodium), then proceeding to the Essentiality section of the search form and asking for genes with ‘Any evidence of essentiality in any species’. Finally, in the same page, under Druggability, one can further restrict the search by requesting only genes with ‘Associated compounds: Curated’. Such a query would produce a list of 39 genes. The user can then obtain a list of the compounds associated with these genes by clicking on ‘Show curated compounds’ from the results page.
THE TDR TARGETS CURATION EFFORT
A target-based drug discovery process is guided by the incremental gathering of data about a target, usually with the final goal of validating the target. Key data about the target's essentiality for the parasite, its expression in a relevant life cycle stage, and its chemical tractability are all available in the literature, and can be extracted and integrated in the database using controlled vocabularies (ontologies) that facilitate querying and cross-relation to other database objects. As previously described (1), the TDR Targets team has compiled extensive literature data on phenotypic responses of targets or whole pathogens to genetic or chemical (pharmacological) perturbations. These literature data nicely complement the genome-scale databasets in providing additional target validation-related information on individual targets or groups of targets that have been the subject of more focused research. For chemicals, our structured representation of the effects of compounds as phenotypes (e.g. decreased cell growth, abnormal morphology, inhibition of catalytic activity, etc.) is distinct from others’ (e.g. ChEMBL's) reporting of these effects as ‘activities’ (e.g. IC50's, % growth inhibition, etc.), which we have also imported into our database. These inconsistencies lead to certain challenges in querying the curated data (see ‘Caveats’ below). Nevertheless, it makes sense to combine our own curated datasets with others because there is relatively little overlap among them. For example, of the ∼450 pathogen targets we have curated internally for association with compounds, only ∼20% have also been curated in the ChEMBL dataset, which is larger but focused mostly on non-infectious diseases. This limited overlap is explained in part by the different journals used in these curation efforts; ChEMBL draws mostly from medicinal chemistry journals such as the Journal of Medicinal Chemistry and Bioorganic & Medicinal Chemistry Letters, while TDR Targets draws heavily from pathogen-specific journals like Molecular and Biochemical Parasitology and Antimicrobial Agents and Chemotherapy. The curatorial work of the TDR Targets team is an ongoing effort and in the future will include inputs solicited from the tropical infectious disease community.
In the current release of TDR Targets, we incorporated data on curated validation credentials for Schistosoma mansoni and Trypanosoma cruzi, therefore completing curation for six key tropical disease causing organisms (species targeted for curation in previous releases of TDR Targets include: Mycobacterium tuberculosis, Trypanosoma brucei, Leishmania major and P. falciparum). The new data contains descriptions of 303 phenotypes (associated with 143 targets and 322 compounds), derived from genetic experiments (for 39 targets) and chemical experiments (for 113 targets). These data complement the data from previous releases for other diseases/organisms, bringing the number of targets with curated validation credentials to 448; and the number of compounds curated internally to 968.
Additionally, we have recently integrated genome-wide phenotyping data for T. brucei derived from the work of Alsford et al. (31). These data, covering different life cycle stages of the parasite, and from different culture conditions, contributed genetic validation information for ∼7400 (80%) of the annotated protein coding genes in the genome, in the form of ‘loss of fitness’ or ‘gain of fitness’ phenotypes. The integrated phenotype descriptions derived from this work are a significant addition to TDR Targets. These correspond to ∼30 000 annotations, about half of all phenotype annotations previously available in the database. These annotations are now searchable and readily available for target prioritizations. Together with gene knockout datasets for M. tuberculosis integrated previously in TDR Targets, they represent the only two examples of genome-wide genetic validation data for WHO target organisms, which are the focus of TDR Targets.
INCORPORATION OF DATA TO ASSESS ASSAYABILITY OF TARGETS
For the purposes of this database, a target is considered assayable if it is an enzyme included in Sigma-Aldrich's collection of assays, or if it has been assayed according to the BRENDA database (32). The BRENDA database contains categories for cloned and purified genes but not assayed genes per se, so to create our ‘assayed’ category we combined entries from the Km and Specific Activity categories, which give the clearest picture of whether a protein has actually been enzymatically assayed. The mapping of the BRENDA entries to genes in TDR Targets was carried out as follows: (i) Mapping by Enzyme Commission (EC) number: EC numbers in BRENDA were used to map the entries to those TDR genes with identical EC numbers; (ii) for BRENDA entries where there was no match to a pathogen gene by EC, the gene was identified by name in the species-specific database (e.g. PlasmoDB) and mapped to that gene; (iii) if there was no gene in the species-specific database with the same EC or name as in the BRENDA entry, the gene was identified by sequence similarity using the sequence from the associated BRENDA literature reference as the query. For each TDR species all entries for the genus were mapped; for example, genes that were assayed/purified/cloned in Plasmodium knowlesi were mapped to P. falciparum and Plasmodium vivax. The source species for the data is specified in the ‘Assayability’ section on the corresponding gene pages.
Aside from having a convenient readout of activity, another aspect of assayability is being able to produce and purify a recombinant form of the protein in question. The largest-scale attempt to express enzymes from TDR Targets species has come through the Structural Genomics of Pathogenic Protozoa (SGPP) and Medical Structural Genomics of Pathogenic Protozoa (MSGPP) project at the University of Washington. Therefore each protein that has been successfully expressed in recombinant form by (M)SGPP is annotated as such in the database, and links are provided for users to access additional information available at these resources (progress in obtaining diffracting crystals, availability of plasmid clones, etc.)
OTHER DATA UPDATES IN TDR TARGETS: NEW GENOMES
Since its launch in 2007 (1), and through several releases, the database has integrated a number of additional genomes. Most important amongst recent additions was the incorporation of helminth genomes (Brugia malayi, a nematode, the causative agent of Filariasis; and Schistosoma mansoni, a trematode flatworm that causes Schistosomiasis). Data from these organisms, including essential annotation were derived from GenBank (5), SchistoDB (33) and GeneDB (3). Other genomes of interest for those studying parasites are T. gondii, the causative agent of Toxoplasmosis, sometimes used as a model organism for studying some aspects of apicomplexan biology that are more difficult to study in e.g. malaria parasites; and P. vivax, an important human malaria parasite which cannot be yet cultured and studied under laboratory conditions. These genomes have also been integrated in TDR Targets, allowing the full range of operations to be performed on their targets.
In addition to integrating the genomes of various pathogenic organisms, TDR targets also includes the genomes of various phylogenetically useful species from which different kinds of datasets can be mapped. The genomes of vertebrates (human, mouse), plants (Arabidopsis thaliana, Oryza sativa) invertebrates (Drosophila melanogaster), nematodes (Caenorhabditis elegans) and closely related species of pathogens already represented in TDR Targets (e.g. Leishmania braziliensis, infantum, and mexicana). These complete proteomes are part of the OrthoMCL database of orthologue groups (7), and allow users to formulate questions such as: ‘search for L. major proteins that are/are not present in L. braziliensis’, or ‘find P. falciparum proteins that are also present in plants or bacteria’ (e.g. when looking for apicoplast associated targets).
CAVEATS: THINGS TO LOOK OUT FOR WHEN SEARCHING TDR TARGETS
A number of important clarifications can be made to help users of the database. These are related to the way chemical information has been curated and integrated in the database. Knowing about these issues will help users make sense of search results, and understand why compounds fail to appear in a result set against all expectations. First, when curating the literature, if a compound has been assayed against an organism (e.g. for inhibition of growth of Leishmania amastigotes), this will be recorded by a curator, even if the activity/phenotype is nil (e.g. no growth inhibition). Therefore when searching the database for compounds with ‘any activity’ a user is actually searching for ‘compounds with any information about their activity’, as the query will return both active and inactive compounds. Second, the activities of compounds are recorded in different forms and units, as specified by the original authors in published papers. Even when a significant effort is invested to standardize the information in the database (e.g. as done by the ChEMBL team), the number of different ways in which activity is reported prevents the formulation of simple filtering queries such as ‘show me all compounds with activity <5 uM’. This type of query can be easily applied to activities reported as IC50s, but does not make sense for activities reported as ‘% inhibition’ (see Figure 3 for a limited list of available activity types). In the latter case, the concentration of the compound used to assess inhibition is usually attached to the assay description (e.g. ‘% inhibition at 1 ug/ml’), and is therefore difficult to query separately. Finally, it is also worthwhile to note here that when substructure or similarity searches are run using a very minimal chemical scaffolds or a very ubiquitous chemical fragment, the number of results retrieved can be enormous and therefore the database currently limits these types of searches.
CONCLUSION
The TDR Targets Database (http://tdrtargets.org) is an online open-access resource developed for the purpose of facilitating target prioritization using a comprehensive collection of data describing gene function, structure, essentiality, assayability and druggability. An extensive set of informatic tools facilitates mining of this resource. The current focus of the database is on pathogenic organisms that are causative agents of tropical infectious diseases as prioritized by the World Health Organization's Special Programme for Research and Training in Tropical Diseases (TDR) and include M. leprae, M. tuberculosis, P. falciparum, P. vivax, S. mansoni, T. gondii, T. brucei, T. cruzi, L. major, and B. malayi and its endosymbiont Wolbachia. Starting with release 4 of the TDR Targets database, chemical information has been integrated into TDR Targets along with cheminformatic tools to run chemical searches taking advantage of a variety of data describing chemical properties and 2D structure of small molecules. These new developments provide a chemogenomics platform that allows users to query the available chemical data, and investigate associations between targets and compounds. The web interface implemented in TDR Targets allows users to seamlessly move from a list of target genes to list of compounds known to interact with these genes and vice versa. As pointed to above, several different databases are now available for browsing and searching chemical datasets and drug targets, but each of them focuses on only a subset of the functionalities available in TDR Targets, and almost none of them is focused on tropical diseases. TDR Targets, therefore, addresses a need that is somewhat neglected by comparable databases and resources. Initially developed to integrate in one place data from parasitic genomes, evaluation of gene function, essentiality and suitability for drug development of targets, TDR Targets now has extended coverage of another key component of the drug discovery process: chemical data including assays, activities, and associations with targets.
A number of key improvements are necessary to keep TDR Targets useful, up to date and relevant for the community of scientists working on tropical diseases. Development of web services and other computational tools to facilitate reuse of data is one area that will be a major focus in the future. Incorporating information on the commercial availability of compounds, and providing links to providers is another key aspect that will be incorporated in future releases. But more importantly perhaps, a sustained curation effort is also required to keep valuable target validation data and compound activity data up to date, and to identify valuable medicinal chemistry data for integration in TDR Targets’ chemical database. As mentioned above, the focus of the TDR Targets curation effort has been largely put on the gathering of information on validating credentials for targets. However, now that a substantial investment has been made into the integration of compound data, curation should be extended to gather other supporting information, such as data on assays, and on the reported activities of compounds (in the form of IC50s, %inhibition, phenotypes, etc.) Adequate funding is much needed to sustain these activities.
FUNDING
Funding for open access charge: Agencia Nacional de Promocion Cientifica y Tecnologica (ANPCyT, Argentina) PICT-2010-1479.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We would like to thank John Overington (EBI) for providing an early release of the Starlite/ChEMBL database for integration into TDR Targets, and Peter Ertl (Novartis) for providing the JME applet for structure-based searches. This work was supported by the Special Programme for Research and Training in Tropical Diseases (TDR), and in part by a grant from the National Agency for the Promotion of Science and Technology (ANPCyT, Argentina, PICT-2010-1479). FA and SJC are fellows of the National Research Council (CONICET, Argentina). MPM is supported by fellowship from a Fogarty International Research Collaboration Award, NIH (FIRCA Grant Number D43TW007888).
REFERENCES
- 1.Agüero F, Al-Lazikani B, Aslett M, Berriman M, Buckner FS, Campbell RK, Carmona S, Carruthers IM, Chan AWE, Chen F, et al. Genomic-scale prioritization of drug targets: the TDR Targets database. Nat. Rev. Drug Discov. 2008;7:900–907. doi: 10.1038/nrd2684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aurrecoechea C, Brestelli J, Brunk BP, Fischer S, Gajria B, Gao X, Gingle A, Grant G, Harb OS, Heiges M, et al. EuPathDB: a portal to eukaryotic pathogen databases. Nucleic Acids Res. 2010;38:D415–D419. doi: 10.1093/nar/gkp941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hertz-Fowler C, Peacock CS, Wood V, Aslett M, Kerhornou A, Mooney P, Tivey A, Berriman M, Hall N, Rutherford K, et al. GeneDB: a resource for prokaryotic and eukaryotic organisms. Nucleic Acids Res. 2004;32:D339–D343. doi: 10.1093/nar/gkh007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lew JM, Kapopoulou A, Jones LM, Cole ST. TubercuList - 10 years after. Tuberculosis. 2011;91:1–7. doi: 10.1016/j.tube.2010.09.008. [DOI] [PubMed] [Google Scholar]
- 5.Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW. GenBank. Nucleic Acids Res. 2011;39:D32–D37. doi: 10.1093/nar/gkq1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jones L, Moszer I, Cole ST. Leproma: a Mycobacterium leprae genome browser. Lepr. Rev. 2001;72:470–477. [PubMed] [Google Scholar]
- 7.Chen F, Mackey AJ, Stoeckert CJ, Roos DS. OrthoMCL-DB: querying a comprehensive multi-species collection of ortholog groups. Nucleic Acids Res. 2006;34:D363–D368. doi: 10.1093/nar/gkj123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berman H, Henrick K, Nakamura H, Markley JL. The worldwide Protein Data Bank (wwPDB): ensuring a single, uniform archive of PDB data. Nucleic Acids Res. 2007;35:D301–D303. doi: 10.1093/nar/gkl971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pieper U, Webb BM, Barkan DT, Schneidman-Duhovny D, Schlessinger A, Braberg H, Yang Z, Meng EC, Pettersen EF, Huang CC, et al. ModBase, a database of annotated comparative protein structure models, and associated resources. Nucleic Acids Res. 2011;39:D465–D474. doi: 10.1093/nar/gkq1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gaulton A, Overington JP. Role of open chemical data in aiding drug discovery and design. Future Med. Chem. 2010;2:903–907. doi: 10.4155/fmc.10.191. [DOI] [PubMed] [Google Scholar]
- 11.Gamo F, Sanz LM, Vidal J, Cozar CD, Alvarez E, Lavandera J, Vanderwall DE, Green DVS, Kumar V, Hasan S, et al. Thousands of chemical starting points for antimalarial lead identification. Nature. 2010;465:305–310. doi: 10.1038/nature09107. [DOI] [PubMed] [Google Scholar]
- 12.Guiguemde WA, Shelat AA, Bouck D, Duffy S, Crowther GJ, Davis PH, Smithson DC, Connelly M, Clark J, Zhu F, et al. Chemical genetics of Plasmodium falciparum. Nature. 2010;465:311–315. doi: 10.1038/nature09099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Seiler KP, George GA, Happ MP, Bodycombe NE, Carrinski HA, Norton S, Brudz S, Sullivan JP, Muhlich J, Serrano M, et al. ChemBank: a small-molecule screening and cheminformatics resource database. Nucleic Acids Res. 2008;36:D351–D359. doi: 10.1093/nar/gkm843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Matos PD, Alcántara R, Dekker A, Ennis M, Hastings J, Haug K, Spiteri I, Turner S, Steinbeck C. Chemical Entities of Biological Interest: an update. Nucleic Acids Res. 2010;38:D249–D254. doi: 10.1093/nar/gkp886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Knox C, Law V, Jewison T, Liu P, Ly S, Frolkis A, Pon A, Banco K, Mak C, Neveu V, et al. DrugBank 3.0: a comprehensive resource for ‘omics’ research on drugs. Nucleic Acids Res. 2011;39:D1035–D1041. doi: 10.1093/nar/gkq1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu T, Lin Y, Wen X, Jorissen RN, Gilson MK. BindingDB: a web-accessible database of experimentally determined protein-ligand binding affinities. Nucleic Acids Res. 2007;35:D198–D201. doi: 10.1093/nar/gkl999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goede A, Dunkel M, Mester N, Frommel C, Preissner R. SuperDrug: a conformational drug database. Bioinformatics. 2005;21:1751–1753. doi: 10.1093/bioinformatics/bti295. [DOI] [PubMed] [Google Scholar]
- 18.Wang G, Li X, Wang Z. APD2: the updated antimicrobial peptide database and its application in peptide design. Nucleic Acids Res. 2009;37:D933–D937. doi: 10.1093/nar/gkn823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barh D, Kumar A, Misra AN. Genomic Target Database (GTD): a database of potential targets in human pathogenic bacteria. Bioinformation. 2010;4:50–51. doi: 10.6026/97320630004050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gao Z, Li H, Zhang H, Liu X, Kang L, Luo X, Zhu W, Chen K, Wang X, Jiang H. PDTD: a web-accessible protein database for drug target identification. BMC Bioinformatics. 2008;9:104. doi: 10.1186/1471-2105-9-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhu F, Han B, Kumar P, Liu X, Ma X, Wei X, Huang L, Guo Y, Han L, Zheng C, et al. Update of TTD: Therapeutic Target Database. Nucleic Acids Res. 2010;38:D787–D791. doi: 10.1093/nar/gkp1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bost F, Jacobs RT, Kowalczyk P. Informatics for neglected diseases collaborations. Curr. Op. Drug Dis. Dev. 2010;13:286–296. [PubMed] [Google Scholar]
- 23.Gaulton A, Bellis LJ, Bento PA, Chambers J, Davies M, Hersey A, Light Y, McGlinchey S, Michalovich D, Al-Lazikani B, et al. ChEMBL: a large-scale bioactivity database for drug discovery. Nucleic Acids Res. 2012;40:D1100–D1107. doi: 10.1093/nar/gkr777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang Y, Xiao J, Suzek TO, Zhang J, Wang J, Bryant SH. PubChem: a public information system for analyzing bioactivities of small molecules. Nucleic Acids Res. 2009;37:W623–W633. doi: 10.1093/nar/gkp456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nwaka S, Hudson A. Innovative lead discovery strategies for tropical diseases. Nat. Rev. Drug Discov. 2006;5:941–955. doi: 10.1038/nrd2144. [DOI] [PubMed] [Google Scholar]
- 26.Oduor RO, Ojo KK, Williams GP, Bertelli F, Mills J, Maes L, Pryde DC, Parkinson T, Van Voorhis WC, Holler TP. Trypanosoma brucei glycogen synthase kinase-3, a target for anti-trypanosomal drug development: a public–private partnership to identify novel leads. PLoS Negl. Trop. Dis. 2011;5:e1017. doi: 10.1371/journal.pntd.0001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tatusov RL, Fedorova ND, Jackson JD, Jacobs AR, Kiryutin B, Koonin EV, Krylov DM, Mazumder R, Mekhedov SL, Nikolskaya AN, et al. The COG database: an updated version includes eukaryotes. BMC Bioinformatics. 2003;4:41. doi: 10.1186/1471-2105-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Willett P. Similarity searching using 2D structural fingerprints. Methods Mol. Biol. 2011;672:133–158. doi: 10.1007/978-1-60761-839-3_5. [DOI] [PubMed] [Google Scholar]
- 29.Haider N. Functionality Pattern Matching as an Efficient Complementary Structure/Reaction Search Tool: an Open-Source Approach. Molecules. 2010;15:5079–5092. doi: 10.3390/molecules15085079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pence HE, Williams A. ChemSpider: an online chemical information resource. J. Chem. Ed. 2010;87:1123–1124. [Google Scholar]
- 31.Alsford S, Turner DJ, Obado SO, Sanchez-Flores A, Glover L, Berriman M, Hertz-Fowler C, Horn D. High-throughput phenotyping using parallel sequencing of RNA interference targets in the African trypanosome. Genome Res. 2011;21:915–924. doi: 10.1101/gr.115089.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Scheer M, Grote A, Chang A, Schomburg I, Munaretto C, Rother M, Söhngen C, Stelzer M, Thiele J, Schomburg D. BRENDA, the enzyme information system in 2011. Nucleic Acids Res. 2011;39:D670–D676. doi: 10.1093/nar/gkq1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zerlotini A, Heiges M, Wang H, Moraes RL, Dominitini AJ, Ruiz JC, Kissinger JC, Oliveira G. SchistoDB: a Schistosoma mansoni genome resource. Nucleic Acids Res. 2009;37:D579–D582. doi: 10.1093/nar/gkn681. [DOI] [PMC free article] [PubMed] [Google Scholar]