Abstract
Mounting evidence suggests that malignant tumors are initiated and maintained by a subpopulation of cancerous cells with biological properties similar to those of normal stem cells. However, descriptions of stem-like gene and pathway signatures in cancers are inconsistent across experimental systems. Driven by a need to improve our understanding of molecular processes that are common and unique across cancer stem cells (CSCs), we have developed the Stem Cell Discovery Engine (SCDE)—an online database of curated CSC experiments coupled to the Galaxy analytical framework. The SCDE allows users to consistently describe, share and compare CSC data at the gene and pathway level. Our initial focus has been on carefully curating tissue and cancer stem cell-related experiments from blood, intestine and brain to create a high quality resource containing 53 public studies and 1098 assays. The experimental information is captured and stored in the multi-omics Investigation/Study/Assay (ISA-Tab) format and can be queried in the data repository. A linked Galaxy framework provides a comprehensive, flexible environment populated with novel tools for gene list comparisons against molecular signatures in GeneSigDB and MSigDB, curated experiments in the SCDE and pathways in WikiPathways. The SCDE is available at http://discovery.hsci.harvard.edu.
INTRODUCTION
Cells in adult non-germinal tissues such as blood, skin and intestine turn over briskly and are known to require stem cells for lifelong renewal. These tissue stem cells are capable of proliferation and self-renewal, and can produce differentiated progeny through the expression of tissue-specific genes. Recent evidence suggests that studying adult stem cells can provide insight into cancer cell biology. Only small fractions of tumor-derived cells are clonogenic in culture or tumorigenic in vivo (1,2). Cancers are therefore thought to rely on the activity of stem or stem-like cells that are tumorigenic and exhibit the cardinal properties of self-renewal and multi-lineage differentiation potential.
Stem and differentiated cells within a tumor are reported to differ in sensitivity toward therapy (3). Studies have independently established embryonic stem cell gene expression signatures where cancer subtypes with poor survival prognosis are enriched in treatment-resistant, stem-like cells. Stem cell signatures resulting in poor prognosis have so far been found in glioma, breast, lung, colon and esophageal cancers (4–10). Comparing stem cell populations therefore has the potential to identify new molecular targets for drug and immune therapies that destroy the self-renewing cancer stem cells (CSCs). However, descriptions of gene and pathway stem-like signatures across cancers are inconsistent across platforms, tissues and laboratories.
Driven by a need to understand CSC molecular profiles generated at the Harvard Stem Cell Institute (HSCI), we have developed a platform to integrate CSC experimental information: the Stem Cell Discovery Engine (http://discovery.hsci.harvard.edu). We have collected, curated and integrated this data into the Stem Cell Discovery Engine (SCDE) to permit molecular comparisons between normal and cancerous stem cells, between stem-cell compartments in blood, intestine and brain, and between mouse models and human tissues.
SCDE overview
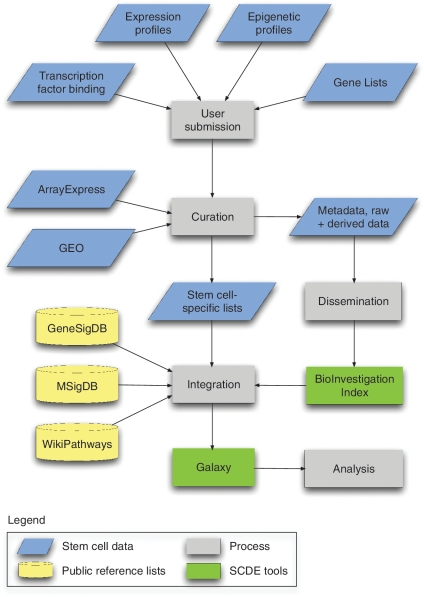
The SCDE is a modular online system designed to handle data submission, curation, analysis, integration and dissemination of stem cell-related experiments (Figure 1). The system has two components: (i) a tissue and cancer stem cell database accessible through the BioInvestigation Index (BII) (11) and (ii) a customized instance of the Galaxy analysis engine (12,13). It includes tools that integrate public stem cell data with user-submitted experiments. Its initial focus is on gene list manipulation, and interaction with the curated Gene Signatures Database (GeneSigDB) (14), Molecular Signatures Database (MSigDB) (15), and WikiPathways pathway database (16) (Figure 1). A description of the database in accordance with BioDBCore standards (17) is available in Supplementary Table S1.
Figure 1.
System architecture diagram showing integration of data into the SCDE BioInvestigation Index (BII) and Galaxy instances. CSC-related experiments are submitted by stem cell researchers or selected from public repositories. After curation using the ISA tools and conversion to ISA-Tab format, the associated metadata, raw data files and processed gene lists are stored in the BII. The stem cell-specific gene lists are transformed into standardized gene identifiers to facilitate integration and comparison against similarly formatted reference lists (GeneSigDB, MSigDB, WikiPathways and other SCDE experiments) within Galaxy.
Curation of experimental metadata and derived data
The SCDE database provides a source of structured experimental information on assays, derived gene lists and pathway profiles. Heterogeneity in experimental information has been reduced by rigorous, manual curation of the experimental model, cell and tissue types, disease state, surface markers and other relevant data. Submitted user data is first checked for relevance, i.e. studies must be performed using well-defined stem cell, tissue stem cell and/or cancer stem cell populations, and must produce genome-scale data with potential to provide insight into the stem-like characteristics of cancers. All of the raw data with its sample characteristics must be available. Data input fields are then mapped to the ontologies listed in Table 1 according to species-specificity and overall coverage of the ontology. New terms are submitted to the ontology maintainers for future inclusion. This ensures that new terms are standardized and incorporated for community use. Experimental protocols and analytical methods are annotated with the goal of providing sufficient information to reproduce or perform similar experiments and to derive the processed data. Derived data in the form of gene lists are converted to standardized identifiers to be used for gene list comparisons within Galaxy.
Table 1.
Curated metadata
| Field | Ontologies (in order of preference) |
|---|---|
| Organism | NEWT UniProt Taxonomy Database (Newt), NCBI Taxonomy (NCBITaxon) |
| Strain | Experimental Factor Ontology (EFO) |
| Developmental stage | EFO |
| Disease state | ICD-9, NCI Thesaurus, Disease Ontology |
| Organism part (tissue type) | Foundational Model of Anatomy, Mouse Gross Anatomy, BRENDA tissue/enzyme source (BTO), EFO |
| Cell type | Cell Type Ontology (CL), EFO |
| Cell line | EFO, NCI Thesaurus |
| Genotype | Ontology for Biomedical Investigations (OBI; depending on species) |
| Cell surface marker | Currently annotated as ±, high/lo. An appropriate standard needs to be developed. |
| Immunoprecipitation antibody | Protein Name (specify manufacturer where available) |
| Binding site | SO (sequence ontology) for methylation sites |
| Phenotypic quality | PATO: Phenotypic qualities (properties) |
| Treatment (perturbation) | PATO, CHEBI: Chemical Entities of Biological Interest OBI (to describe perturbations such as genetic modification, transient expression) |
We stored experimental metadata in the Investigation/Study/Assay (ISA-Tab) format, i.e. high level information about the experiment is recorded in the ‘investigation’ file, sample attributes and factors in the ‘study’ files, and protocols and analysis methods in the ‘assay’ files. This general purpose tab-delimited grammar manages metadata from diverse studies, and enables users to align with community-defined minimum information, ontologies and checklists (11,18,19). It comes with support tools for curation (including semi-automated annotation tagging through the NCBO BioPortal annotation service (20) to speed the process) and format conversion (http://isatab.sourceforge.net) to make it straightforward to submit data to international public repositories, such as the Gene Expression Omnibus (GEO) (21). ISA-Tab is supported and maintained by a global collaboration of biocurators (22). While the initial cost of curation is high, it allows for sharing of ISA-Tab configurations we have developed specifically for stem cell data that can be used within the various ISA tools by the stem cell community. The goal is to build a curation network and establish community involvement so that standards are agreed upon and adopted.
Database contents
A primary focus was a selection of studies related to normal and CSCs, and in particular for three model systems—blood, intestine and brain. In these tissues, the behavior of native stem cells is especially well characterized, investigators generally agree on stem cell definitions, and cancer is common. Table 2 shows the distribution of data across organisms, tissues and types of measurements. The database integrates 53 public studies comprised of 1098 molecular assays from CSC-related experiments from multiple tissues, species and heterogeneous platforms. Five additional studies comprised of 84 assays are stored as private, unpublished data that are available to specific researchers upon login and are ready for dissemination upon publication. Fifteen studies were contributed by researchers in the HSCI community and an additional 40 studies related to CSC biology were selected from StemBase (23,24). Forty-six studies were performed in rodent models and 13 in human cells; these include two studies containing samples assayed from both rodent and human models. The database is made up in large part by microarray expression profiling studies but results from nucleotide sequencing (i.e. ChIP-seq) studies of histone methylation and transcription factor binding, histology and expression analysis by RT–PCR are also included.
Table 2.
SCDE data
| Studiesa | Assaysa | |
|---|---|---|
| Organism | ||
| Mouse | 45 (4) | 846 (54) |
| Human | 13 (1) | 255 (30) |
| Rat | 1 | 18 |
| Tissue type | ||
| Blood/bone marrow | 20 (3) | 374 (48) |
| Muscle | 8 | 125 |
| Brain/neural | 6 (1) | 68 (6) |
| Intestine | 4 | 39 |
| Mammary | 2 | 34 |
| Skin | 2 (1) | 135 (30) |
| Measurementb | ||
| Transcription profiling | 57 (5) | 1161 (84) |
| Histone modification profiling | 2 | 42 |
| Transcription factor binding site identification | 1 | 21 |
| Tissue histology | 1 (1) | 6 (6) |
aTotal number of studies and assays; the number of private studies and assays are shown in brackets
bFurther information and details of technology platforms are available online at http://discovery.hsci.harvard.edu.
Data acquisition and dissemination
Researchers can submit their own data or suggest public data to a curator, who manually curates it according to community-accepted standards and ontologies (Table 1). In cases where published studies have associated data deposited in ArrayExpress, the MAGEtoISA converter tool permits rapid conversion from MAGE-TAB to ISA-Tab format, which is then manually evaluated by a curator for completeness and corrected where necessary.
To ensure that all stem cell data are comparable, primary and derived data sets are organized in a standardized manner and disseminated to the public using a local instance of the SCDE Bioinvestigation Index (BII). This data repository is designed to support storage, querying and display of multi-omics data sets (11). The annotated metadata allows users to search the entire corpus of experiments in the BII based on organism, measurement type (e.g. transcriptional profiling), technology (e.g. nucleotide sequencing), and platform (e.g. Illumina) or to search free text across all fields (Figure 2A). Study pages display the details of each experiment (Figure 2B–D). The annotation has focused on ensuring that cell types, tissues and experimental variables are consistently reported to improve query capabilities, and to establish sound annotation practices to describe stem cell research (e.g. descriptions of genetic modifications).
Figure 2.
Screenshots showing elements of the BioInvestigation Index browse view. (A) The results of a free text search using the term ‘intestine’ that retrieves four studies—two human and two murine—that include transcription profiling using DNA microarrays, and ChIP-seq for transcription factor binding and histone modifications; (B) One matching record, SHIVDASANI-S-1, with descriptive text; (C) Annotated experimental factor metadata including time, antibodies used in the chromatin immunoprecipitation (ChIP) experiments, and cell differentiation status; (D) Annotated sample metadata including the tissue and cell types, using the Foundational Model of Anatomy (FMA) and Brenda Tissue (BTO) ontologies, respectively; (E) Download panel for the raw and processed data.
Published studies are automatically made publicly available. ISA-Tab formatted metadata can be downloaded for information pertaining to the assays, such as normalization procedures for microarray experiments and GEO accession identifiers where available. Raw primary data (e.g. CEL files for Affymetrix microarrays) and processed derived data (e.g. author-generated gene lists) can also be downloaded from the BII using the ‘Raw Data’ and ‘Processed Data’ buttons (Figure 2E). Alternatively, the data can be accessed within the SCDE Galaxy framework for analysis as described in the following section. Researchers with the appropriate access permissions can query unpublished data to perform early analyses, and upon publication, have the added benefit of exporting their ISA-Tab formatted data for submission to ArrayExpress using the conversion tools. The corresponding functionality for submission to GEO in MiniML format is in progress and will represent a valuable incentive for the stem cell community to use the SCDE as a first port of call for submission of CSC functional genomics data.
Querying CSC molecular signatures using Galaxy
In addition to querying experimental metadata, the SCDE provides functionality to interrogate stem cell molecular profiles in a linked Galaxy instance, with the goal of identifying similarities and differences between normal and cancer stem cell experiments. All raw and processed data stored in the BII and several additional manually curated stem cell-related gene lists are accessible from within Galaxy for analysis.
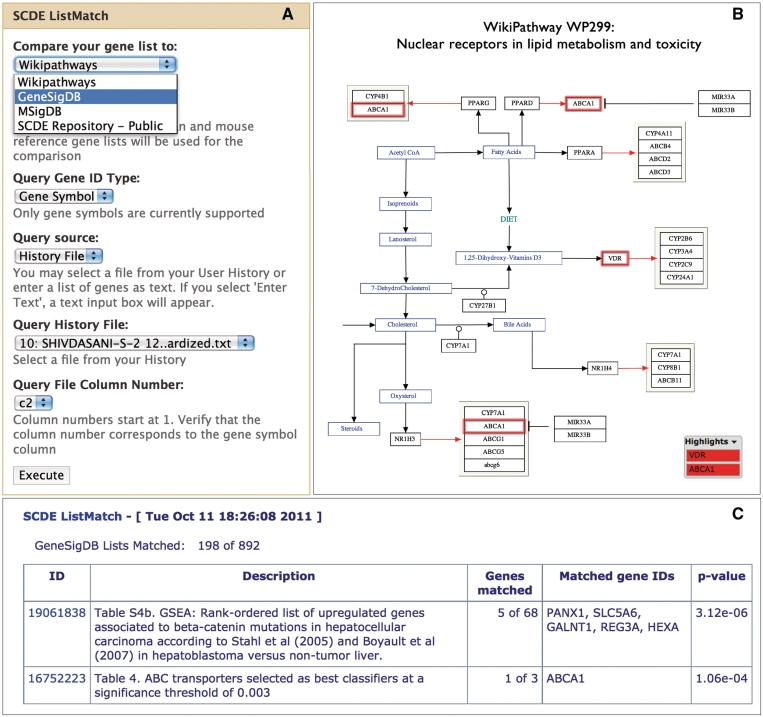
Manual curation and consistent identifier conversion differentiate the SCDE from other gene list comparison tools. Derived gene lists have been mapped to standardized gene symbols using methods developed for GeneSigDB. Such standardization allows for comparisons to determine genes that are shared or unique across experiments. Tools are available to compare a single gene list (SCDE ListMatch) or multiple gene lists (SCDE ListCompare) against curated gene signatures in GeneSigDB, molecular signatures in MSigDB, derived gene lists from the SCDE database and pathways in WikiPathways. These tools allow users to identify genes in common with defined reference signatures and pathways (Figure 3). Results are summarized and ranked according to a hypergeometric test P-value and linked to the relevant overlapping gene sets (Figure 3B). For WikiPathways comparisons, a link is provided to visualize the gene matches in color-coded diagrams of canonical pathways (Figure 3C). The SCDE Intersect tool identifies genes that are common to multiple gene lists. By using the Galaxy interface, users can maintain a record of their analysis history and easily compare multiple data sets stored in their history.
Figure 3.
Composite figure showing the results of a ListMatch query using the set of intestinal differentiation genes that are reduced upon Cdx2 depletion from the SHIVDASANI-S-2 study. (A) SCDE ListMatch input page with options to compare against WikiPathways, GeneSigDB, MSigDB and the SCDE repository. (B) Results of the query against WikiPathways projected onto the canonical pathway representation with matching genes highlighted in red (partial screenshot shown). (C) Querying against GeneSigDB results in a top match to genes related to liver cancer.
DISCUSSION
The SCDE database provides a repository for curated CSC data and a framework for developing methods to compare molecular information on stem cell related populations. We illustrate the functionality of the SCDE using the following use case as an example. A leukemia researcher enters the SCDE through the BII interface. A search for the term ‘leukemia’ in the free text search box produces five transcriptional profiling studies performed in mouse models. The user selects the first result (ARMSTRONG-S-1) to obtain further details of the study and is provided with information about genetic modifications, hematopoietic progenitor cell types, immunophenotypes, type of leukemia studied and the mouse strain used in the experiment. Wishing to perform a related experiment, the researcher downloads the experimental metadata in ISA-Tab format, which provides him with additional information about the sample cell types, labeling protocol, microarray chip used, number of replicates, normalization procedure, etc. After performing his experiment, the researcher returns to the SCDE to determine how similar his results are to the ARMSTRONG-S-1 study, or indeed to any of the experiments in the SCDE. Using the Galaxy web interface, he uploads his list of differentially expressed genes from his leukemia experiment and uses the ListMatch tool to determine the following: (i) significant overlap with gene signatures from SCDE experiments (this may reveal similarities to the leukemia studies or other hematopoietic stem cell experiments contained in the SCDE; (ii) genes enriched in curated signatures from GeneSigDB or mSigDB (such overlaps provide information about similar diseases states, positional biases and functional groupings) and (iii) genes that overlap with known pathways from WikiPathways (genes are projected onto the canonical pathway diagram to indicate where they occur within the pathway). Going a step further, the researcher uses the ListCompare tool to find the overlap with the ARMSTRONG-S-1 gene list with reference to canonical pathways in WikiPathways. This allows him to identify pathways that contain genes from both lists even where the intersection of the two lists is small, generating hypotheses about possible pathways to study further. Having done his analysis within Galaxy, the researcher saves the gene lists, parameters and results and can share this data with his collaborators or make it publicly available.
The SCDE is unique in its community-oriented approach for identifying relevant experiments, capturing and curating study information, and integrating new analysis capabilities compared to previous resources. The adoption of the ISA-Tab format permits inclusion of multiple diverse data types and demonstrates that the tools we have used are ready for scale up. The Galaxy framework allows us to rapidly add relevant analysis methods developed by the growing Galaxy development community in which we are active participants. The implementation of open source software projects that are gaining community support will ensure that the SCDE continues to evolve. The tools developed for the SCDE Galaxy instance have been published on bitbucket at the URL http://bitbucket.org/hbc/galaxy-central-hbc. By publishing this resource and making the infrastructure available, we hope to develop the stem cell community and obtain feedback on annotation practices, relevant data sets and analytical methods.
Future directions
While the comparison of gene signatures is informative, a systematic approach to compare and determine the role of key pathway contributions across different experimental systems and cancers against a consistent background is needed. A pathway fingerprinting method to determine functional similarity among experiments independently of platform or species is being developed for integration into the SCDE (Altschuler et al., submitted). We will continue to expand the SCDE to include additional CSC-related studies and new data types, and work with the stem cell community to further refine relevant ontology terms, as has been the case for the Cell Ontology (25). A further focus will be to develop methods to integrate epigenetic data with gene expression. Scientists interested in adding or curating studies, or in implementing analysis options that are not yet available, are encouraged to contact us at scde@hsci.harvard.edu.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health (1RC2CA148222-01 to R.S., S.A. and W.H.); Harvard Stem Cell Institute. Funding for open access charge: Harvard School of Public Health Dean's Fund; National Institutes of Health Stimulus awards.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We acknowledge assistance from the Harvard Stem Cell Institute for contributing CSC data and are grateful for discussions and assistance from Dr Amit Sinha. We thank Dr Miguel Andrade-Navarro for access to the StemBase data. Thank you also to Emily Merrill and Dr Sudeshna Das for their input on ontology usage.
REFERENCES
- 1.Dick JE. Stem cell concepts renew cancer research. Blood. 2008;112:4793–4807. doi: 10.1182/blood-2008-08-077941. [DOI] [PubMed] [Google Scholar]
- 2.Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001;414:105–111. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- 3.Bao S, Wu Q, McLendon RE, Hao Y, Shi Q, Hjelmeland AB, Dewhirst MW, Bigner DD, Rich JN. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006;444:756–760. doi: 10.1038/nature05236. [DOI] [PubMed] [Google Scholar]
- 4.Ben-Porath I, Thomson MW, Carey VJ, Ge R, Bell GW, Regev A, Weinberg RA. An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat. Genet. 2008;40:499–507. doi: 10.1038/ng.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kappadakunnel M, Eskin A, Dong J, Nelson SF, Mischel PS, Liau LM, Ngheimphu P, Lai A, Cloughesy TF, Goldin J, et al. Stem cell associated gene expression in glioblastoma multiforme: relationship to survival and the subventricular zone. J. Neurooncol. 2010;96:359–367. doi: 10.1007/s11060-009-9983-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Onaitis M, D'Amico TA, Clark CP, Guinney J, Harpole DH, Rawlins EL. A 10-gene progenitor cell signature predicts poor prognosis in lung adenocarcinoma. Ann. Thorac. Surg. 2011;91:1046–1050. doi: 10.1016/j.athoracsur.2010.12.054. discussion 1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pece S, Tosoni D, Confalonieri S, Mazzarol G, Vecchi M, Ronzoni S, Bernard L, Viale G, Pelicci PG, Di Fiore PP. Biological and molecular heterogeneity of breast cancers correlates with their cancer stem cell content. Cell. 2010;140:62–73. doi: 10.1016/j.cell.2009.12.007. [DOI] [PubMed] [Google Scholar]
- 8.Varnat F, Duquet A, Malerba M, Zbinden M, Mas C, Gervaz P, Ruiz i Altaba A. Human colon cancer epithelial cells harbour active HEDGEHOG-GLI signalling that is essential for tumour growth, recurrence, metastasis and stem cell survival and expansion. EMBO Mol. Med. 2009;1:338–351. doi: 10.1002/emmm.200900039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sjolund J, Manetopoulos C, Stockhausen MT, Axelson H. The Notch pathway in cancer: differentiation gone awry. Eur. J. Cancer. 2005;41:2620–2629. doi: 10.1016/j.ejca.2005.06.025. [DOI] [PubMed] [Google Scholar]
- 10.Yang L, Bian Y, Huang S, Ma X, Zhang C, Su X, Chen ZJ, Xie J, Zhang H. Identification of signature genes for detecting hedgehog pathway activation in esophageal cancer. Pathol. Oncol. Res. 2011;17:387–391. doi: 10.1007/s12253-010-9337-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rocca-Serra P, Brandizi M, Maguire E, Sklyar N, Taylor C, Begley K, Field D, Harris S, Hide W, Hofmann O, et al. ISA software suite: supporting standards-compliant experimental annotation and enabling curation at the community level. Bioinformatics. 2010;26:2354–2356. doi: 10.1093/bioinformatics/btq415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Blankenberg D, Von Kuster G, Coraor N, Ananda G, Lazarus R, Mangan M, Nekrutenko A, Taylor J. Galaxy: a web-based genome analysis tool for experimentalists. Curr. Protoc. Mol. Biol. 2010;Chapter 19:Unit 19 10 11–21. doi: 10.1002/0471142727.mb1910s89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goecks J, Nekrutenko A, Taylor J. Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol. 2010;11:R86. doi: 10.1186/gb-2010-11-8-r86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Culhane AC, Schwarzl T, Sultana R, Picard KC, Picard SC, Lu TH, Franklin KR, French SJ, Papenhausen G, Correll M, et al. GeneSigDB–a curated database of gene expression signatures. Nucleic Acids Res. 2010;38:D716–725. doi: 10.1093/nar/gkp1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liberzon A, Subramanian A, Pinchback R, Thorvaldsdottir H, Tamayo P, Mesirov JP. Molecular signatures database (MSigDB) 3.0. Bioinformatics. 2011;27:1739–1740. doi: 10.1093/bioinformatics/btr260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pico AR, Kelder T, van Iersel MP, Hanspers K, Conklin BR, Evelo C. WikiPathways: pathway editing for the people. PLoS Biol. 2008;6:e184. doi: 10.1371/journal.pbio.0060184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gaudet P, Bairoch A, Field D, Sansone SA, Taylor C, Attwood TK, Bateman A, Blake JA, Bult CJ, Cherry JM, et al. Towards BioDBcore: a community-defined information specification for biological databases. Database. 2011;2011:baq027. doi: 10.1093/database/baq027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brazma A. Minimum Information About a Microarray Experiment (MIAME)–successes, failures, challenges. ScientificWorldJournal. 2009;9:420–423. doi: 10.1100/tsw.2009.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sansone SA, Rocca-Serra P, Field D, Maguire E, Taylor C, Hide W, Hofmann O, Fang H, Neumann S, Tong T, et al. Towards interoperable bioscience data. Nature Genetics. 2011 doi: 10.1038/ng.1054. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Whetzel PL, Noy NF, Shah NH, Alexander PR, Nyulas C, Tudorache T, Musen MA. BioPortal: enhanced functionality via new Web services from the National Center for Biomedical Ontology to access and use ontologies in software applications. Nucleic Acids Res. 2011;39:W541–W545. doi: 10.1093/nar/gkr469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barrett T, Troup DB, Wilhite SE, Ledoux P, Rudnev D, Evangelista C, Kim IF, Soboleva A, Tomashevsky M, Marshall KA, et al. NCBI GEO: archive for high-throughput functional genomic data. Nucleic Acids Res. 2009;37:D885–D890. doi: 10.1093/nar/gkn764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sansone SA, Rocca-Serra P, Brandizi M, Brazma A, Field D, Fostel J, Garrow AG, Gilbert J, Goodsaid F, Hardy N, et al. The first RSBI (ISA-TAB) workshop: “can a simple format work for complex studies?”. Omics. 2008;12:143–149. doi: 10.1089/omi.2008.0019. [DOI] [PubMed] [Google Scholar]
- 23.Porter CJ, Palidwor GA, Sandie R, Krzyzanowski PM, Muro EM, Perez-Iratxeta C, Andrade-Navarro MA. StemBase: a resource for the analysis of stem cell gene expression data. Methods Mol. Biol. 2007;407:137–148. doi: 10.1007/978-1-59745-536-7_11. [DOI] [PubMed] [Google Scholar]
- 24.Sandie R, Palidwor GA, Huska MR, Porter CJ, Krzyzanowski PM, Muro EM, Perez-Iratxeta C, Andrade-Navarro MA. Recent developments in StemBase: a tool to study gene expression in human and murine stem cells. BMC Res. Notes. 2009;2:39. doi: 10.1186/1756-0500-2-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bard J, Rhee SY, Ashburner M. An ontology for cell types. Genome Biol. 2005;6:R21. doi: 10.1186/gb-2005-6-2-r21. [DOI] [PMC free article] [PubMed] [Google Scholar]