Abstract
The mapper2 Database (http://genome.ufl.edu/mapperdb) is a component of mapper2, a web-based system for the analysis of transcription factor binding sites in multiple genomes. The database contains predicted binding sites identified in the promoters of all human, mouse and Drosophila genes using 1017 probabilistic models representing over 600 different transcription factors. In this article we outline the current contents of the database and we describe its web-based user interface in detail. We then discuss ongoing work to extend the database contents to experimental data and to add analysis capabilities. Finally, we provide information about recent improvements to the hardware and software platform that mapper2 is based on.
INTRODUCTION
mapper2 is a web-based platform for the analysis of transcription factor binding sites (tfbss) in multiple genomes. Its development was motivated by the need to provide a complete and comprehensive platform for the computational analysis of tfbss, an essential task for the investigation of genetic regulatory networks at the genome-wide scale (1–3). mapper2 is composed of four interconnected modules: the ‘Database’, the ‘Search Engine’, ‘rsnps’, and the ‘Wizard’.
In this article we describe the improvements and extensions to the database that have been performed since its original publication (4). After an overview of the architecture of the mapper platform, we focus on the mapper2 Database, describing work performed on the database itself, work performed on the web-based user interface to increase its usability and effectiveness, and other miscellaneous features including hardware and software improvements.
SYSTEM ARCHITECTURE
Model libraries
mapper2 is a modular, web-based system for the computational analysis of tfbss. At its core is a library of >1000 models, each associated with a specific transcription factor (tf). A model is a computational representation of the known binding sites for a tf, that can be used to detect the presence of putative binding sites in an arbitrary dna sequence. In most cases, the sites recognized by a tf are degenerate, and cannot be represented by a simple consensus sequence (5). Therefore, modeling tfbss in general requires a probabilistic representation of the distribution of nucleotides in the site. The most common of these representations is the position weight matrix (pwm), whose contents describe the observed frequencies of the four nucleotides at each position in the alignment of all known binding sites (6).
In mapper2, tfbs models are represented by ‘hidden Markov models’, and are generated using the hmmer package (version 2) (7). hmmer provides commands to ‘train’ and ‘optimize’ models (called HMMs in the following), and to scan a dna sequence using one or more models in order to detect sites that match the nucleotide pattern described by them (henceforth called ‘hits’). Each hit receives a numerical score representing the likelihood that the dna sequence at the position where the hit was found matches the nucleotide distribution encoded by the model. Using HMMs to represent binding sites offers advantages in terms of specificity and sensitivity over PWMs, and provide more flexibility in modeling sites with complex structures (such as sites composed of two fragments separated by a spacer) (8).
mapper2 includes three large libraries of tfbs models. The first one (transfac models) was built using the same optimal alignments used to build the PWMs in the transfac database (9). The second one (mapper2 models) is specific to mapper2, and was built using tfbs sequences from transfac that were not used in the transfac models. The third library was generated using tfbs data from the jaspar database (10). Table 1 summarizes the number of models in each library and the TFs they represent. It should be noted that there may be multiple models for the same tf, in case the source databases contain different sets of binding sites for it. mapper2 provides a table listing all models in its libraries (with links to pages providing details about each one), that can be sorted either by model accession or by factor name.
Table 1.
Number of hmm models and factors represented in each of the three default model libraries
| Library | Models | Factors |
|---|---|---|
| TRANSFAC | 399 | 326 |
| MAPPER | 529 | 434 |
| JASPAR | 89 | 89 |
| Total | 1017 | 678 |
Modules
The mapper2 database includes all putative binding sites detected in the upstream regions of all known human, mouse and Drosophila transcripts using all the models in the mapper2 libraries. For each transcript, the region analyzed extends from 10 000 bp upstream of the transcription start site to 50 bp downstream of the translation start site (ATG), in order to include the proximal promoter, possible upstream regulatory elements and initial untranslated exons. Multiple transcripts produced by the same gene are analyzed independently.
Information stored in the database for each hit includes its exact genomic position, the model that generated it, the gene in whose promoter it was found, the hit alignment (showing the model's representative sequence and the matched dna sequence) and the predictive score assigned to it by hmmer. Table 2 summarizes the contents of the mapper2 database, showing the number of sequences analyzed and the total number of hits in each genome.
Table 2.
Summary of the contents of the mapper2 database, by organism
| Human | Mouse | Drosophila | |
|---|---|---|---|
| Models | 832 | 829 | 819 |
| Genes | 21 510 | 21 736 | 14 003 |
| Promoters | 34 022 | 27 443 | 22 369 |
| Total bases (Mb) | 628 | 578.6 | 266 |
| Total hits | 33 122 746 | 24 313 246 | 19 248 318 |
| Hits/promoter | 973.5 | 886.0 | 860.5 |
| Hits/model | 1.17 | 1.06 | 1.05 |
| Hits spacing | 18.95 | 23.80 | (13.82) |
‘Models’ indicates the number of models that produced at least one hit in a genome. ‘Hits/model’ represents the average number of hits in a promoter for each model. ‘Hits spacing’ is the average number of nucleotides between hits (see text for an explanation of the Drosophila value).
The mapper2 search engine can be used to scan an arbitrary dna sequence in real time. While the database includes hits found only in the upstream regions of known genes, the search engine interface allows the user to specify the exact region of each known gene to be scanned, or to upload one or more sequences in FASTA format. Although slower than querying the database, running a search engine analysis provides higher flexibility in selecting the sequences to be analyzed, and is not limited to the three default genomes.
The rsnps module is used to identify single-nucleotide polymorphisms (SNPs) with potential regulatory effects. This is accomplished by detecting tfbss containing SNPs, and measuring the change in score due to the allelic change. Score changes above a set threshold indicate that the snp has the potential to disrupt the binding site, reducing its binding affinity for the factor or deleting it altogether.
The mapper2 wizard is a tool allowing the user to create new hmm models, by uploading a multiple-sequence alignment of binding sites. The wizard generates the hmm, optimizes it and stores it in the user's private library. The private model can then be used in the search engine or in rsnps in addition to the ones from the default libraries.
mapper2 modules are tightly interconnected: they are all based on the same model libraries and genomic annotation database, and they present a consistent user interface for ease of use. The user can therefore freely switch between them, executing multiple analysis session in parallel. Sessions are run in the background, and the system provides a page through which the user can display all running sessions, with a progress indicator showing the percentage of the analysis completed, and all completed ones (allowing to download previously generated results).
DATABASE
Genomes and annotations
mapper2 now uses the most recent versions of the human, mouse and Drosophila genomes and annotations. In particular, it uses assembly GRCh37/hg19 of the human genome, NCBI37/mm9 of the mouse genome and BGDP Release 5/dm3 of the Drosophila genome. Genomic annotations (e.g. gene names and identifiers) are kept up to date by an automated procedure, and the whole database is automatically re-created when new releases of the genomes or of the source binding site databases become available.
Binding site data
The current version of the mapper2 database was built using binding site data from transfac version 11.2 and the most recent version of jaspar (October 2009). Table 1 reports the number of models and represented factors in each of the three default libraries.
After generating the models, we collected the dna sequences for the upstream regions of all known genes in the three genomes known to mapper2, and we scanned them with all models. The resulting hits were stored in a relational database, that serves as the main data source for mapper2. Table 2 reports the number of promoters scanned and hits detected in the three genomes. The increase in the number of hits compared to the previous version of the database is mainly due to the higher number of known transcripts now available for the three organisms.
Note that the value of the density of hits in Drosophila is misleading, due to the small size of this genome. Since the region analyzed for each gene is at least 10 000 bp long, the total amount of sequence scanned is larger than the size of the entire genome. Moreover, in many cases the scanned region for a gene will overlap other close genes; it is therefore advisable to select a small region upstream of the transcript start when working with Drosophila.
According to Table 2, every model is represented at least once in each promoter, which is not biologically realistic. This is actually an artifact of the hmmer algorithm, that always produces at least one hit per sequence scanned (albeit with a very low score). A more meaningful analysis can be performed by selecting only those sites with a score at or above a specific threshold, in order to reduce the number of false positives. The threshold can be selected on the basis of the distribution of scores in the database for each model, for example choosing a desired percentile level. The mapper2 interface provides for filtering hits at various percentiles levels ranging from 50th to 99th; in our experience, percentile levels of 90 or above should be used in a real-world analysis.
An alternative way of defining a high-quality threshold consists in scanning a random sequence (having the same average base composition of typical gene promoters), generating the distribution of scores for the resulting hits and selecting an extreme value from it (for example, the 99th percentile). This translates into a 1% chance that a hit with a score above this value occurred by chance, and is therefore a false positive.
Table 3 reports the results of the randomization procedure just described. For each organism, we generated a 5 Mbp random sequence with a nucleotide distribution derived from a large number of gene promoters, and we scanned it with each model, computing the 99th percentile of the resulting distribution of scores. This value was used as the high-quality threshold for the hits contained in the database for that model. The table displays the total number of high-quality hits in each organism, and the associated statistics.
Table 3.
Summary of hits with scores above a threshold corresponding to a false discovery rate of 1%, obtained by scanning a 5 Mb randomized promoter sequence
| Human | Mouse | Drosophila | |
|---|---|---|---|
| High quality hits | 12 354 505 | 8 849 004 | 6 828 654 |
| Hits/promoter | 363.1 | 322.5 | 305.2 |
| Hits/model | 0.44 | 0.49 | 0.37 |
| Hits distance | 50.8 | 65.39 | (39.0) |
The number of models and of promoters analyzed is the same as in Table 2.
USER INTERFACE
The mapper2 user interface has been extensively redesigned and improved in order to increase its usability and effectiveness. The main design principles behind it are ‘simplicity’ (all input fields are documented, with examples) and ‘consistency’ (all modules use a similar interface and display results using the same output layout).
A menu bar at the top of each page provides access to all mapper2 functions and utilities. Commands are divided into five sections: ‘MAPPER’ (containing general commands such as Register, to create a private account, and News); ‘Models’ (with links to the list of all mapper2 models and to the mapper2 wizard); ‘Tools’ (containing links to the three main mapper2 modules); ‘Private’ (only available when logged in, containing links to the user's mapper2 sessions and private models); ‘Support’ (providing the Help and Feedback commands, and a link to a page listing publications about mapper2).
Inputs
A mapper2 database session is initiated by clicking on the Database link in the main menu. This leads to a form used to input all necessary parameters, that is divided into three sections: ‘Gene(s)’, ‘Organisms’ and ‘Models’.
The Gene(s) section provides controls to specify which gene or genes the user is interested in. There are four options, selected using a radio button. To start, the user may enter one or more gene identifiers in a text field; mapper2 automatically recognizes a large number of standard gene identifiers including official gene symbols, gene IDs, mrna accession numbers, ensembl identifiers. In case the user specifies a gene that has multiple isoforms, all of them will be analyzed at the same time. Alternatively, the user may upload a list of identifiers from a file. The behavior of the system in this case is the same, but this option is more practical when dealing with a large number of genes. The third option allows the user to select a pathway from a menu, to automatically analyze all the genes it contains. This option is useful when looking for TFs that may act as common regulators for several genes in the same pathway. Pathway definitions are taken from the KEGG database (11).
When using one of these three options, the user can also select the size of the upstream region of each gene to be analyzed, as a number of base pairs upstream of the transcript start or the ATG. It should be noted that all hits in the region up to 10 000 bp upstream of the transcript start are precomputed and stored in the database; these controls simply determine the number of hits that will be returned to the user.
Finally, the fourth option is to perform a ‘models-only’ search: in this case, the user does not specify any gene but selects one or more models in the Models section (see below), and the system will return the top-scoring hits for those models in the entire genome.
The Organisms section serves two purposes. The first one is to indicate the genome the user is interested in, for these cases in which this cannot be determined from the inputs. For example, mrna accession numbers are organism-specific, while gene names are not (there may be genes in different organisms with the same name). Therefore, when using gene names or when performing a models-only search, it becomes necessary to indicate the genome of interest. The second part of this section allows the user to perform searches based on homology by selecting additional organisms. If one or more of the genes being analyzed has a homolog in the selected organisms, the homologs will be automatically added to the session.
The Models section is used to specify which models to retrieve hits for. The user can select one or more of the three standard libraries, or enter a list of individual model identifiers. This last option is facilitated by a pop-up window listing all available models. The list can be sorted either by model accession number or by factor name; clicking on an accession number adds the corresponding model to the list.
When the input parameters have been set, the user should proceed by clicking the button at the bottom of the form. The system will then display a summary page recapitulating the inputs, and the user has the choice to go back to the input page (in case anything needs to be modified) or to start the analysis. While the analysis is running, the system displays a progress indicator and a link that can be used to retrieve the results when ready. Since mapper2 sessions run in the background, the user is free to go on using the system to start other sessions, or to leave the site. When the session is complete, the user will receive a notification by email; alternatively, the status of the session can be checked in the ‘My Hitsets’ page at any time.
Results
The page displaying analysis results has undergone a total redesign, aimed at making it more useful and effective. The page is divided into two sections: a panel at the top providing information or controls, and a table listing all hits. The contents of the panel are divided into five different subpages, that can be selected using the tabs at the top: ‘Summary’, ‘Inputs’, ‘Filters’, ‘Display’, and ‘Export’.
The ‘Summary’ page contains general information about the session being displayed, starting with its identifier, a brief description, its status (that will normally be ‘Done’) and the total run time. The next line (‘Displayed run’) contains a menu allowing the user to select the run to be displayed, in case the session contains multiple runs (e.g. when analyzing all genes in a pathway). The ‘Displayed sites’ item shows the number of hits displayed in the table, out of the total number of hits produced.
The ‘Inputs’ page provides information about the parameters of the run being displayed, including details about the gene (symbol, name, mrna accession, organism), the region of the promoter that was scanned and the model libraries used in the analysis.
The ‘Filters’ page provides commands to change the set of displayed hits. Hits can be selected based on their score, either by entering a threshold value (so that only hits with a score above the threshold are displayed) or by selecting a percentile level from a menu. Since different models have different score distributions, this option allows the user to select high-quality hits independently of the actual numerical values of their scores. This page also allows selecting hits based on the name of the factor they apply to, and highlighting hits in evolutionarily conserved regions.
The ‘Display’ page contains options to control how hits are displayed in the main table. Currently, they consist of a menu to select how hit coordinates are displayed (absolute on chromosome, relative to transcript start, relative to ATG), and one to select how hits are sorted (by position, score, E-value, factor name or factor accession).
The ‘Export’ page provides controls to export the hits in a variety of different ways. To start, the user may choose the export format among the following:
‘text’ — a delimited file with one row for each hit;
‘alignments’ — similar to text, but including the hit alignment;
‘BED’ — suitable for upload to the UCSC Genome Browser (12) as a custom track;
‘GFF’ — the General Feature Format defined by the Wellcome Trust Sanger Institute.
‘image’ — a graphical representation of the analyzed region showing the position and factor name for all hits.
The user can then select the delimiter to use when generating files in ‘text’ or ‘alignments’ format (tab or comma), and the name of the file (by default, a randomly generated session identifier). Two further controls are available in the ‘Options’ section. When the ‘compress’ checkbox is active, the generated file will be compressed with gzip for faster downloads. When ‘all results’ is checked, mapper2 will produce a file containing hits for all runs in the current session, instead of those of the currently displayed run only. Finally, the user can choose how to receive the exported hits: by downloading a file (using the ‘Export’ button), or by email (using the ‘Email’ button). The page also provides a link to automatically upload the results to the UCSC genome browser and to display them as a custom track.
The hits table displays all hits that are visible according to the settings in the ‘Display’ section. If the number of hits is over 100, the table is initially hidden, and can be displayed by clicking a button. When the table is visible, it contains 12 columns, showing:
gene symbol, entrez identifier and transcript accession number;
accession number of the model that produced the hit and factor name;
strand, chromosome, start and end position of the hit;
hit score and E-value.
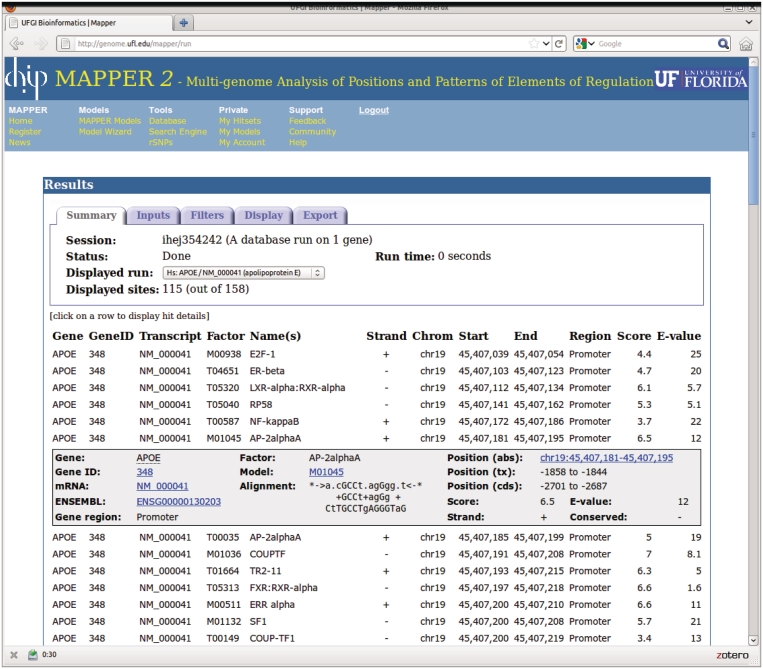
Clicking the mouse button over a row opens a box containing more information about the hit in that row. Additional fields displayed in this case include the ensembl gene identifier, the hit alignment, the hit position according to all three reference systems (absolute on chromosome, relative to transcript start, relative to ATG) and a flag indicating whether the hit lies in an evolutionarily conserved region. Moreover, several fields in this box are hyperlinks to pages with further information. For example, the Gene ID is linked to the ncbi Gene page for that gene; the model accession number is a link to the mapper2 page describing that model, and the absolute hit position is a link to the UCSC Genome Browser. Figure 1 shows a typical results page for a single-gene database run.
Figure 1.
The mapper2 page displaying results of a single-gene database query. The gray box shows detailed information for the hit in the line directly above it.
MISCELLANEOUS
Interoperability
mapper2 provides a way for external programs to perform database queries without going through the web-based interface. Requests are submitted in the form of a special url that encodes the search parameters (gene identifiers, model accession numbers, score threshold, etc.), and the results are returned as a tab-delimited file in Alignments format (see the description of the Export command). This mechanism is similar to the one adopted by the ncbi Entrez website to implement its EUtils interface (13). Although it does not provide all the functionality of a true ‘Web-Services’ interface, it is extremely easy to implement and use; mapper2 searches can therefore easily be incorporated in automated annotation and analysis pipelines. A description of the available search options and of the way in which the request should be formatted can be obtained by accessing the db-rpc interface at the url http://genome.ufl.edu/mapper/db-rpc.
Hardware and software platforms
The mapper2 database has been moved to a more powerful server at the University of Florida (16-core GNU/Linux machine, 48 GB RAM), and is now directly accessible at the url http://genome.ufl.edu/mapperdb/. Its previous url (http://mapper.chip.org/) is still accessible, but will automatically redirect to the new location.
mapper2 is written in Common Lisp, a high-performance object-oriented language ideally suited for complex applications (14). In addition to upgrading to the 64bit version of the language, we have adopted a package providing ‘persistence’ for internal application objects. This means that mapper2 sessions, including input data and result sets, are permanently stored on the server (until the user who generated them decides to delete them) in an automated and secure way. Considering that a result set may contain thousands of hits, this solution is more efficient and reliable than storing them in a separate, external database system, while still providing protection against loss of data due to server crashes.
Future work
Future plans for the development of the mapper2 database include adding more genomes, and updating the primary binding site data to a more recent version of transfac. We are also going to investigate the inclusion of ChIP-Seq data in the database, in order to show which predicted tfbss are in agreement with experimental data.
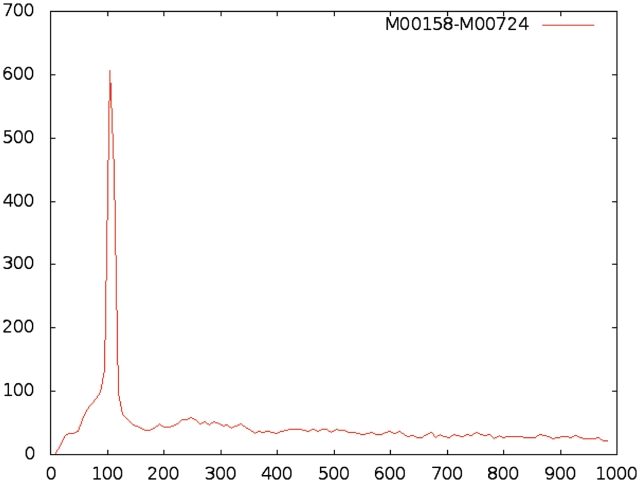
In addition, we are performing an analysis on the spatial distribution of binding sites and on the co-occurrence of pairs of binding sites. In the first case, we are interested in determining whether a tf preferentially binds at a specific distance upstream of the transcription start site, or if instead its binding sites are uniformly distributed over the promoter. In the second case, we consider each possible pair of models and we determine the distribution of the distances between their hits; if the distribution exhibits one or more peaks, this indicates that the two factors preferentially bind at specific distances from each other. We will perform these analyses on all models in the database, and make the results accessible through the system's interface. These data will help in the interpretation of search results and in the formulation of biological hypotheses based on the spatial arrangement of tfbss. Figure 2 shows the distance distribution plot for an example pair of binding sites.
Figure 2.
Example distance distribution plot for a pair of models. The graph represents the histogram of distances between binding sites for models M00158 (HNF-4) and M00724 (HNF-3α) in mouse. In the vast majority of cases, these binding sites are separated by 100bp, while other distances occur at very low and almost constant frequencies.
AVAILABILITY
mapper2 is freely available to any user. Users are encouraged to create individual accounts, but the system can be used in ‘guest’ mode without any loss of functionality. Accounts provide users with the ability to store generated result sets and private models in a secure area of the website. The contents of the database are available for download as tab-delimited files by request.
CONCLUSIONS
The new version of mapper2 described in this article represents a significant improvement over the initial one described in Ref. (4). The contents of the database are constantly being updated as new genome releases and new binding site data become available. The user interface has been completely redesigned for usability and consistency with the rest of the mapper2 platform, with which the Database module is now totally integrated. An http-based interface allows other programs to query the mapper2 database, that can therefore become a component in a distributed annotation and analysis pipeline.
Future work will be aimed at incorporating experimental data information, such as ChIP-Seq data, to assist in the validation and interpretation of the computational prediction provided by the system. We are also developing methods to analyze the spatial distribution of binding sites in promoters, and the relative distances of pairs of binding sites in order to study interactions between synergistic transcription factors.
FUNDING
Funding for open access charge: UF Genetics Institute.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The author wishes to thank Voichita D. Marinescu and Isaac S. Kohane for their collaboration on previous versions of mapper; Eric F. Tsung, Brandon M. Walts, Ettore Rizzo and Eric Hernandez for their contributions to the development of the current version; and the anonymous reviewers for their useful suggestions.
REFERENCES
- 1.Bolouri H, Davidson EH. Modeling transcriptional regulatory networks. Bioessays. 2002;24:1118–1129. doi: 10.1002/bies.10189. [DOI] [PubMed] [Google Scholar]
- 2.Davidson EH, Rast JP, Oliveri P, Ransick A, Calestani C, Yuh CH, Minokawa T, Amore G, Hinman V, Arenas-Mena C, et al. A genomic regulatory network for development. Science. 2002;295:1669–1678. doi: 10.1126/science.1069883. [DOI] [PubMed] [Google Scholar]
- 3.Siggia ED. Computational methods for transcriptional regulation. Curr. Opin. Genet. Dev. 2005;15:214–221. doi: 10.1016/j.gde.2005.02.004. [DOI] [PubMed] [Google Scholar]
- 4.Marinescu VD, Kohane IS, Riva A. The MAPPER database: a multi-genome catalog of putative transcription factor binding sites. Nucleic Acids Res. 2005;33:D91–D97. doi: 10.1093/nar/gki103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Badis G, Berger MF, Philippakis AA, Talukder S, Gehrke AR, Jaeger SA, Chan ET, Metzler G, Vedenko A, Chen X, et al. Diversity and complexity in DNA recognition by transcription factors. Science. 2009;324:1720–1723. doi: 10.1126/science.1162327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kel AE, Gssling E, Reuter I, Cheremushkin E, Kel-Margoulis OV, Wingender E. MATCH: a tool for searching transcription factor binding sites in DNA sequences. Nucleic Acids Res. 2003;31:3576–3579. doi: 10.1093/nar/gkg585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eddy SR. A probabilistic model of local alignment that simplifies statistical significance estimation. PLoS Comput Biol. 2008;4:e1000069. doi: 10.1371/journal.pcbi.1000069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marinescu VD, Kohane IS, Riva A. MAPPER: a search engine for the computational identification of putative transcription factor binding sites in multiple genomes. BMC Bioinformatics. 2005;6:79. doi: 10.1186/1471-2105-6-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wingender E, Chen X, Fricke E, Geffers R, Hehl R, Liebich I, Krull M, Matys V, Michael H, Ohnhuser R, et al. The TRANSFAC system on gene expression regulation. Nucleic Acids Res. 2001;29:281–283. doi: 10.1093/nar/29.1.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sandelin A, Portales-Casamar E, Thongjuea S, Kwon AT, Arenillas D, Zhao XB, Valen E, Yusuf D, Lenhard B, Wasserman WW. JASPAR 2010: the greatly expanded open-access database of transcription factor binding profiles. Nucleic Acids Res. 2010;38:D105–D110. doi: 10.1093/nar/gkp950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kanehisa M, Goto S, Furumichi M, Tanabe M, Hirakawa M. KEGG for representation and analysis of molecular networks involving diseases and drugs. Nucleic Acids Res. 2010;38:D355–D360. doi: 10.1093/nar/gkp896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dreszer TR, Rhead B, Clawson H, Barber GP, Haussler D, Kent WJ, Karolchik D, Kuhn RM, Hinrichs AS, Zweig AS, et al. The UCSC genome browser database: update 2010. Nucleic Acids Res. 2010;38:D613–D619. doi: 10.1093/nar/gkp939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sayers E. E-utilities quick start. 2008 http://www.ncbi.nlm.nih.gov/books/NBK25500/ (17 November 2011, date last accessed) [Google Scholar]
- 14.Seibel P. Practical Common Lisp. NY, USA: Apress; 2005. [Google Scholar]