Abstract
PLEXdb (http://www.plexdb.org), in partnership with community databases, supports comparisons of gene expression across multiple plant and pathogen species, promoting individuals and/or consortia to upload genome-scale data sets to contrast them to previously archived data. These analyses facilitate the interpretation of structure, function and regulation of genes in economically important plants. A list of Gene Atlas experiments highlights data sets that give responses across different developmental stages, conditions and tissues. Tools at PLEXdb allow users to perform complex analyses quickly and easily. The Model Genome Interrogator (MGI) tool supports mapping gene lists onto corresponding genes from model plant organisms, including rice and Arabidopsis. MGI predicts homologies, displays gene structures and supporting information for annotated genes and full-length cDNAs. The gene list-processing wizard guides users through PLEXdb functions for creating, analyzing, annotating and managing gene lists. Users can upload their own lists or create them from the output of PLEXdb tools, and then apply diverse higher level analyses, such as ANOVA and clustering. PLEXdb also provides methods for users to track how gene expression changes across many different experiments using the Gene OscilloScope. This tool can identify interesting expression patterns, such as up-regulation under diverse conditions or checking any gene’s suitability as a steady-state control.
INTRODUCTION
PLEXdb (Plant Expression Database) is a gene expression-based resource to bridge between genotype to phenotype through transcript profiling. PLEXdb integrates multiple data sets from a wide variety of plant and plant pathogen microarrays and provides a single site to access, analyze and disseminate expression data for comprehensive comparative functional genomics studies (1,2). The goal of PLEXdb is to make this data easily accessible to help users answer biological questions and begin to leverage existing results from related large-scale expression studies.
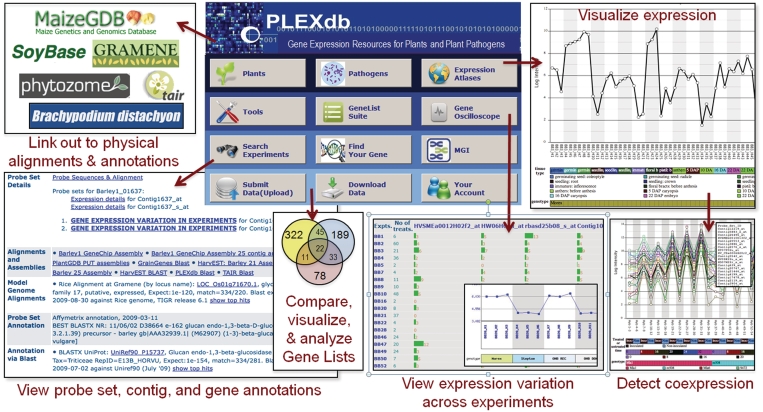
The primary goal of the PLEXdb resource is to provide integration of data and tools that are currently accessible only from disparate resources. Without this integration, researchers, students and teachers would have to download expression data from a repository site; check for conformity to standards that would allow cross-experiment comparisons; map the respective array (or RNA-Seq tags) to genes and those genes to genomic locations and orthologs in other species; install local software for expression data analysis; rely on disparate resources to view associated data and develop their own methods to post-process results, (e.g. obtain additional sequence data for upload into promoter motif-finding software). Thus, PLEXdb facilitates these many different tasks using a single web interface that is easily accessible to any researcher within two or three clicks from the PLEXdb front page (See Fig. 1).
Figure 1.
Interconnection between PLEXdb and a few of its partner databases. Utilizing the PLEXdb annotation portal, every transcript (for every PLEXdb organism) will be presented as an expression profile (upper right), linked to analytical functions (Gene OcilloScope, MPT, MGI, etc.), and aligned on species-specific genome browsers to sequenced and model genomes (e.g., maize, soybean Brachypodium, grape, etc.), providing one-stop-shopping for the plant genome community.
PLEXdb is complementary and synergistic to other expression data archives such as NCBI-GEO and ArrayExpress. General repositories, such as the Gene Expression Omnibus (GEO) (3) and ArrayExpress (4), act as central data distribution hubs for species ranging from E. coli to humans. General repositories make the data available to public users, but because of their large scope and lack of specificity they do not provide community annotation.
The most useful expression databases for on-line analysis and data exploration tend to focus on particular species or problems as they contain links to useful annotation as well as graphics and tools focused at a particular task. Examples of these databases include GENEVESTIGATOR (5) and the Sol Genomics Network (SGN) (6). GENEVESTIGATOR has an extensive set of on-line tools for analysis and visualization of Arabidopsis and other model species microarray data, with searches based on tissue type and developmental stage, but it lacks annotation links and does not allow public submission or download of data. SGN provides a unified resource with sequence, expression and pathway data but it is restricted to a single clade.
DESIGN REQUIREMENTS AND FUNCTIONALITY
The key design requirements for PLEXdb are to allow users to explore data sets and put them into biological context for interpretation. This requires that the database contains carefully annotated and curated experiments with links to controlled vocabularies so that appropriate comparisons can be made for experiments. Gene expression elements must also be thoroughly documented and annotated with timely information. Careful comparisons across species and mapping onto key model organisms allow biologist users to use existing structures to enhance their analyses. Finally, the database must be easy to use and the data should be presented in a form amenable to interpretation. To meet these goals, the PLEXdb team:
Encourages users to submit fully annotated experiments to PLEXdb which meet/exceed the MIAME/Plant requirements (7).
Provides up-to-date annotation of probe sets from a variety of sources such as clade-specific databases, RefSeq, UniPROT and PlantGDB (8).
Creates visualizations that allow users to quickly explore experiment quality and consistency prior to using data in their analyses.
Provides analysis tools such as the Gene List Suite which steps users through the analysis process of creating, analyzing, annotating and managing a gene list.
PLEXdb content
PLEXdb supports all of the available plant and pathogen GeneChip arrays including, Affymetrix 57K Rice, 61K Wheat, 22K Barley1, 18K first-generation Maize, 8K Sugarcane, Fusarium graminearum (9), 61K Soybean/Phytopthora/soybean cyst nematode, 16K Grape, Arabidopsis ATH1 as well as Cotton, Poplar, Citrus, Tomato and Medicago.Recent additions include the newly developed nine-plant pathogenic fungal genome array, as well as the full-genome Brachypodium and Maize arrays. PLEXdb also supports the NimbleGen microarray platforms for Vitis and maize (10). All arrays enjoy complete annotation and analysis support.
Gene atlas experiments have been specially tagged by the PLEXdb curator to highlight experiments that focus on different plant tissues, developmental stages and important experimental conditions such as biotic and abiotic stress. These experiments allow users to quickly see how their gene of interest behaves under different conditions.
The PLEXdb curator reviews the submitted data for overall data quality and looks for signs of common errors such as sample association errors (a sample associated with the wrong data file) using replicate correlation plots. In all cases, the submitter is asked to review the normalized data and is requested to approve the results before posting the data. The curator also regularly reviews GEO for new plant and pathogen array data sets. When high quality new data sets are discovered in GEO (11), the curator determines the experimental factors and imports the data into PLEXdb.
GENE ANNOTATION
Users can find their gene of interest on a specific expression platform via the PLEXdb tool, Find Your Gene, which blasts sequence against the consensus sequence for each microarray.
In addition to linking expression information from experiments to probe sets, PLEXdb provides consensus sequences and annotation for expression elements by linking probe sets to information in several databases, including UniProt, PlantGDB (8) and the Dana Farber Cancer Institute (DFCI) Gene Index (12). It also associates probe set IDs with annotation data from other sources, such as organism-specific consortiums such as Gramene (13), TAIR (14), MaizeGDB (15) and the Fusarium graminearum Database (FGDB) (16). When model genomes are available, annotations and links to alignment tools (e.g. Model Genome Interrogator) are provided. Links to appropriate clade- or organism-specific databases are made (e.g. Gramene for gene models for rice, GrainGenes for physical maps of wheat). The connections to other databases are in most cases determined through BLAST (BLASTN, BLASTX, or TBLASTX). Links to PlantGDB-assembled unique transcripts (PUT) assemblies are also provided for every applicable GeneChip (8).
The expression elements are also linked to gene ontology terms via the UniPROT links. Links to metabolic pathway information allow users to know what pathway their genes are in. Various pipelines exist for the data behind the microarray annotation. For example, the PLEX team runs different types of BLAST and BLASTX against a variety of references bi-annually. The results are stored and used for updating GO and PO tables. These pipelines are all implemented in Perl.
EXPERIMENT ANNOTATION AND PROCESSING
PLEXdb uses the MIAME/Plant (7) guidelines to provide as complete a description of each experiment as possible. Wherever possible, strict structures and controlled vocabularies are used. Of primary importance to enable quick understanding and to facilitate machine-searchable experiments is the use of a factor/level description of the experiment treatment structure. PLEXExpress, the submission tool is used to enable MIAME compliance and use of controlled vocabularies (17). Submitters are also encouraged to include images in their submissions.
PLEXdb uses a factorial design structure that allows for easy comparison between conditions. A factor is a condition that is changed between samples. For example, a factor may be the genotype, type of pathogen inoculation, stress condition, or time point. A level is a specific change in a factor. For example, an experiment might test differences between genotypes A and B. In this case, the factor is ‘genotype’ and its levels are ‘A’ and ‘B’.
Wherever possible, PLEXdb uses the Plant Ontology (18) (PO) terms for development stages, cell types, organism parts and other controlled term lists. Many of these lists come from the MIAME/plant requirements (7); e.g. the terms for describing growth media. This helps pave the way toward comparative expression data analysis and meaningful meta-analyses.
The experiments submitted to PLEXdb may be kept private to the submitter, shared with a group of collaborators only, or made visible to the public. This enables researchers to use PLEXdb as a collaborative tool while a study is ongoing. An experiment submitter can also request a reviewer access code so that reviewers can look at the data from an experiment while evaluating a paper. In accordance with journal policies, upon publication of the primary manuscript, data is considered public.
PLEXdb requires submitted experiments to provide, at a minimum, the raw data files and sample and protocol information. Other file types are optional and all submitted files are available for public downloading when the experiment is made visible to the public. If the researcher requests it, PLEXdb submits the formatted experiments and meta-data to GEO in the name of the researcher and his/her lab. For each experiment, all files provided by the submitter are made available for download according to the visibility of the experiment (private, group or public). In addition, PLEXdb provides the normalized data, tables of treatment means and medians and a tab-separated text file correlating CEL files to treatments and replicates.
After an experiment has been submitted and reviewed by the curator for completeness and correctness, the raw data is normalized by using the Robust Multichip Average (RMA) (19) method and by the Affymetrix MAS5.0 normalization. Several visualizations are generated, including RNA degradation, box plots of raw and normalized intensities by all treatments across the experiment, treatment clustering across the experiment, various treatment scatter plots. This pipeline was constructed using Perl, R, Bioconductor and the Affymetrix Power Tools. For experiments using multi-species GeneChips (Soybean, Medicago), custom probe definition files are used for the RMA normalization step to enable masking out expression elements from species that are not relevant to the experiment.
With improvement in sequencing and sequence assembly technology, there have been significant revisions of the gene model versions available in many species. As a consequence, the current probe configuration of a significant number of probe sets is not congruent with the updated versions of the corresponding gene models. To address this issue, PLEXdb has begun, on a pilot basis, remapping probes as assemblies evolve. For example, the Nimblegen Maize platform is being mapped to the gene models from the maize RefGen V2 build in collaboration with MaizeGDB. Data has been re-normalized based on the new configuration where the gene models serve as the revised probe sets. The new data set has been released as a clone of the original data set which corresponds to the V1 build (10). This approach will make it easier to integrate microarray data with RNAseq/NGS expression data that relies on alignment to the most recent gene models available in a species.
PLEXdb analysis tools
PLEXdb provides a number of tools for submitting, viewing and analyzing experiments, and for creating, and analyzing gene lists. In addition, extensive tutorials have been written on the tools in the database that describes how the tools work with detailed examples.
MODEL GENOME INTERROGATOR
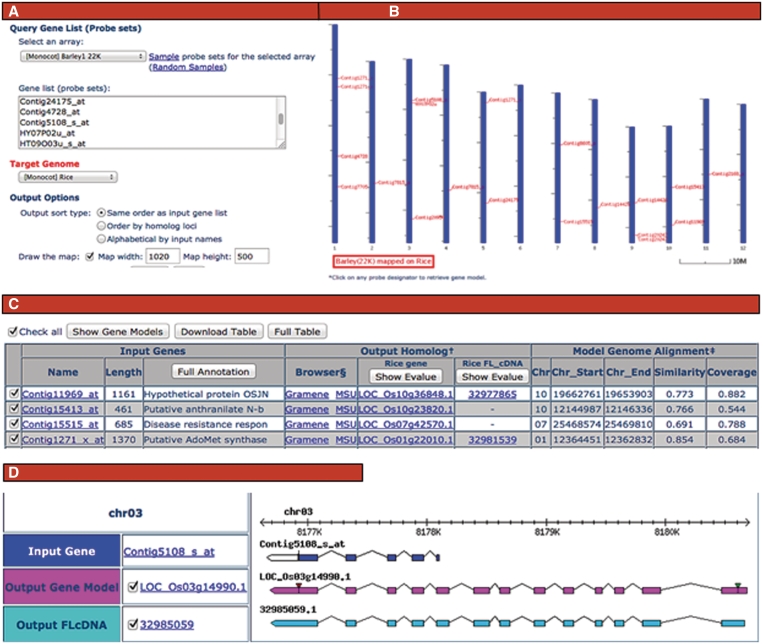
The Model Genome Interrogator (MGI), Version 3 provides structural genomic support for integrated and comparative exploration of gene expression data (2). Based on user input of single or batch queries of microarray probe set identifiers from most of the microarray platforms supported by PLEXdb, MGI uses the sequenced genomes, the annotate protein-coding genes, cDNA and locus coordinate data of either rice or Arabidopsis to identify putative orthologs for the source gene that correspond to the probesets. For each putative ortholog identified, MGI allows researchers to view annotations, visually evaluate gene models and extract sequence data from promoters, exons, introns and UTRs (Figure 2).
Figure 2.
Collage of web interface panels from the PLEXdb implementation of MGI. On the input page (A), users select the source of the genes and the model genome, paste a gene list into the text field, select output preferences and submit the query (20). Map (B) and tabular (C) outputs then display genomic positions and provide detailed annotations with links to PlantGDB and Gramene or TAIR. Users may further evaluate the putative orthologs using the gene model display page (D) (21), where they may also specify the output gene model or FLcDNA which can be viewed at PlantGDB.
On the input page, users must enter a list of gene identifiers or probe set names, specify the expression platform from which they originate, choose the model genome to interrogate and select desired output options. Links to sample data that show format are available on the input page. These are helpful for exploring MGI utilities without requiring prior data analysis.
The output consists of a map of where potential orthologs physically map in the model genome and their identities and annotations. Query probesets that cannot be mapped onto the chosen model genome are listed as ‘Missed Genes’. The GeneSeqer and BLAST results describe the quality of the match and its evidence. The other four columns in the table provide annotations of the query probesets and their matching model gene loci, including direct links to Gramene (13) for rice or TAIR for Arabidopsis (14). More than one row of data is used to summarize the results if more than one locus qualifies as a match. It is common for a query to match multiple annotated protein-coding genes or full length cDNAs associated with a single locus.
The tools allow single or batch extraction of promoter, 5′-UTR, 1st exon, 1st intron and 3′-UTR sequences. The researcher may select the source of evidence to be the annotated protein-coding gene only, or also include full-length cDNA evidence provided by PlantGDB (8).
The 16 microarray platforms mapped onto the rice and Arabidopsis genomes by the PLEXdb implementation of MGI display three levels of connectivity and fidelity that are directly related to phylogenetic distance.
GENE OSCILLOSCOPE
The Gene OscilloScope is a data-mining tool that searches for microarray experiments in PLEXdb where the expression of a queried gene fluctuates (oscillates) the most. Given an expression element, it displays the extent of fluctuation of the treatment means in each of the experiments visible to the user at PLEXdb for the corresponding microarray platform.
Expression of most genes does not change significantly from one treatment to another in any given experiment except those responding to the treatment and are of biological interest. The extent of fluctuation of the expression of a gene in any experiment is measured by the coefficient of variation (CV) of the treatment means for this gene. CV is a measure of the deviation from the mean and is expressed as a percentage from the mean. CV is unaffected by high or low values of the treatment means. As a result, this indicator is not biased towards genes with more transcript abundance. CV tends to underestimate the extent of fluctuation in an experiment with more treatments. For this reason, the Gene OscilloScope tool displays the number of treatments in the table to aid users in making their decision.
The Fluctuation Filter also scans for genes based on their fluctuation in expression measured by the same metric, CV. Given a data set, it searches for all the genes in the data set that have a specified range of CV, e.g. <1% or >25%, etc. It can find genes that show high or low responses to the treatments in a set of experiments. It can find genes that are suitable for steady-state controls in a variety of studies that are less likely to fluctuate under diverse experimental conditions.
GENE LIST SUITE
The gene-list-processing wizard guides users through PLEXdb functions for creating, analyzing, annotating and managing gene lists. Users can upload their own lists or create them from the output of PLEXdb tools, and then apply diverse higher level analyses. The goal is a step-by-step wizard that is easy to use without heavy study and reading of documentation.
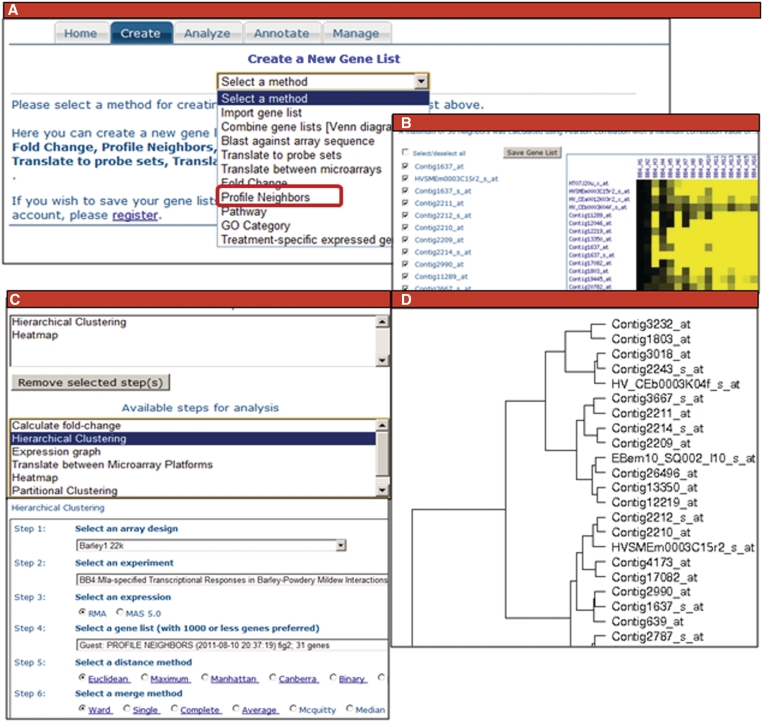
Gene lists can be created using correlated neighbors of a target gene, fold change under different conditions, common Gene Ontology (GO) terms and pathway membership as shown in Figure 3. Gene lists can also be imported from offline analysis or an interesting publication. Set operations such as union and intersection can be carried out on the created lists. Once a gene list is created, the user can analyze the list using clustering, ANOVA, etc. A user can set up a group of analyses to perform on the data set. The analysis itself and the results can be stored for registered users. Analyses for guest users are stored for a limited time only. After analysis, the gene list can be annotated and saved in a spreadsheet.
Figure 3.
Collage of web interface panels from the PLEXdb Gene List Suite. On the input page (A), users select what they would like to do. (B)shows the inputs and outputs for creating a gene list by selecting profile neighbors. Users may further evaluate the gene list using the Analysis page (C), where they may select multiple analysis steps to be used for the gene list. (D) shows the results of hierarchically clustering the gene list of profile neighbors using BB4 data sets.
Applications of PLEXdb
As an example of the utility of PLEXdb, we present a use case.
Map all of the expressed genes associated with a particular condition on the genome
An investigator has completed a series of gene expression measurements, performed statistical analysis, uploaded the data and has a list of genes associated with a particular treatment. The investigator then wishes to see where all genes that are co-regulated or that belong to a particular gene family map on the genome. This is particularly useful for investigators interested in high-throughput quantitative trait (QTL) analysis. For fully annotated sequenced genomes, such as rice or Arabidopsis, Medicago, soybean, poplar and maize, this will be possible by a straightforward look-up of the pre-calculated coordinates in a genome browser. However, the problem is more complex for species without fully sequenced genomes, for example wheat or barley. In this case, it is desirable to identify syntenic positions on the most closely related model genome. These map locations could then be used to search for associations with trait loci to integrate gene expression data with phenotype data. The investigator may also be interested in which gene families and pathways (GO or IUPAC terms) are implicated in the list of co-regulated genes. The investigator may then want to see conserved genes or pathways in another organism (e.g. Arabidopsis) to build hypotheses regarding function by transitive inference and comparison of expression profiles of similar experiments in the other organism.
The integration of data sources, visualizations and analytic tools at PLEXdb facilitate this process. For example, as shown in Figure 2, the Model Genome Interrogator can perform batch mapping of expression elements onto model genomes. This tool allows a user to enter a list of genes derived from any expression experiment (20,21) (A) and immediately visualize their positions on the rice genome (for monocots) or Arabidopsis (for dicots) (B). Using the table that is generated below the map, the user can then get details on position and alignment of orthologous ESTs by a direct link to Gramene or annotation in terms of predicted function.
DEVELOPMENT AND CHALLENGES FOR EXPRESSION DATABASES
There are many challenges faced by expression databases, including new and increasingly prevalent data types, such as RNA-seq, tiling arrays and whole-genome array platforms. Part of the challenge will be finding core identifiers that reach across assemblies and technologies to unify transcriptome data. In addition, probe sets for early array releases were designed several years ago from the available EST assemblies. As a consequence, the current probe configuration of a significant number of probe sets in microarray platforms is no longer congruent with the updated versions of the corresponding gene models. This means that probe sets must be remapped and annotated.
In addition to unifying data across a dizzying array of platforms, the data need to be integrated with databases that provide focused resources for a species or clade to help put transcriptomics data into the proper perspective. Interactive connections with sequence centric, community genome databases provide easy access to physical alignments, genetic map positions and known phenotypes for all genes. The challenge is to make the expression and community data easily accessible for analysis.
For integration of RNA-seq data, new statistical methods to detect how diverse treatments affect alternative promoter use, splicing and other aspects of RNA processing and metabolism will need to be established, as well as meta-analysis methods to facilitate comparison of results across data sets from different experiments, different species and different technologies. This will be especially important for users investigating ‘orphan’ genomes (i.e. no reference transcriptome or genome).
For this to be possible, the data models from both DNA-array and RNA-seq resources must converge. Microarray data has been represented as raw values per probe per biological sample and normalized values (quantile normalization at PLEXdb) after summarization takes the form, per probeset (gene) per sample. For example, a popular representation of RNA-seq data, the Reads Per Kilobase of exon model per Million mapped reads (RPKM) (22), derived from the raw read counts, can be treated as analogous to the raw data from CEL or pair files from Affymetrix or NimbleGen. Quantile normalization can be performed on this data to eventually represent it as an expression value per gene per sample. The best method of normalizing RNA-seq data is still being debated. The availability of new reference genomes will require updated assemblies of archived short-read data, and hence the need for resources to attend to these reiterative efforts.
CONCLUSIONS
Ultimately, omics databases need to fill a community need by providing the biologist user with easy access to analysis capabilities for diverse plant and pathogen transcriptome data sets. PLEXdb seeks to provide a consistent web interface for plant transcriptome data, so that the user can access diverse types of data from multiple starting points, e.g. a particular gene, an experimental factor, physical or genetic map position (i.e. location within a QTL) or gene expression data.
FUNDING
National Science Foundation (DBI-0543441; IOS-0922746); United States Department of Agriculture–Agricultural Research Service Project (3625-21000-049-00D to R.W.). Funding for open access charge: grant funding from NSF.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The PLEXdb team would like to thank the following undergraduate students who have helped develop tools and import experimental data: Jodan Goldie, Royce Blackledge, Jack (Pu) Hou, Joseph Grgic, Emmanuel Owusu, Akul Singhania, Ajani Thomas and Andrew Couch. The PLEXdb team also thanks Ethalinda Cannon of MaizeGDB for her work mapping expression probes to the maize v2 (5b.60) assembly.
REFERENCES
- 1.Shen L, Gong J, Caldo RA, Nettleton D, Cook D, Wise RP, Dickerson JA. BarleyBase--an expression profiling database for plant genomics. Nucleic Acids Res. 2005;33:D614–D618. doi: 10.1093/nar/gki123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wise RP, Caldo RA, Hong L, Shen L, Cannon E, Dickerson JA. BarleyBase/PLEXdb: a unified expression profiling database for plants and plant pathogens. Methods Mol. Biol. 2007;406:347–363. doi: 10.1007/978-1-59745-535-0_17. [DOI] [PubMed] [Google Scholar]
- 3.Barrett T, Troup DB, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH, Sherman PM, et al. NCBI GEO: archive for functional genomics data sets—10 years on. Nucleic Acids Res. 2011;39:D1005–D1010. doi: 10.1093/nar/gkq1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Parkinson H, Sarkans U, Kolesnikov N, Abeygunawardena N, Burdett T, Dylag M, Emam I, Farne A, Hastings E, Holloway E, et al. ArrayExpress update—an archive of microarray and high-throughput sequencing-based functional genomics experiments. Nucleic Acids Res. 2011;39:D1002–D1004. doi: 10.1093/nar/gkq1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hruz T, Laule O, Szabo G, Wessendorp F, Bleuler S, Oertle L, Widmayer P, Gruissem W, Zimmermann P. Genevestigator v3: a reference expression database for the meta-analysis of transcriptomes. Adv Bioinformatics. 2008;2008:420747. doi: 10.1155/2008/420747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bombarely A, Menda N, Tecle IY, Buels RM, Strickler S, Fischer-York T, Pujar A, Leto J, Gosselin J, Mueller LA. The Sol Genomics Network (solgenomics.net): growing tomatoes using Perl. Nucleic Acids Res. 2011;39:D1149–D1155. doi: 10.1093/nar/gkq866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zimmermann P, Schildknecht B, Craigon D, Garcia-Hernandez M, Gruissem W, May S, Mukherjee G, Parkinson H, Rhee S, Wagner U, et al. MIAME/Plant - adding value to plant microarrray experiments. Plant Methods. 2006;2:1. doi: 10.1186/1746-4811-2-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Duvick J, Fu A, Muppirala U, Sabharwal M, Wilkerson MD, Lawrence CJ, Lushbough C, Brendel V. PlantGDB: a resource for comparative plant genomics. Nucleic Acids Res. 2008;36:D959–D965. doi: 10.1093/nar/gkm1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lysøe E, Seong KY, Kistler HC. The transcriptome of Fusarium graminearum during the infection of wheat. MPMI. 2011;24:995–1000. doi: 10.1094/MPMI-02-11-0038. [DOI] [PubMed] [Google Scholar]
- 10.Sekhon R, Lin H, Childs K, Hansey C, Buell RC, de Leon N, Kaeppler S. Genome-wide atlas of transcription through maize development. Plant J. 2011;66:553–563. doi: 10.1111/j.1365-313X.2011.04527.x. [DOI] [PubMed] [Google Scholar]
- 11.Barrett T, Troup DB, Wilhite SE, Ledoux P, Rudnev D, Evangelista C, Kim IF, Soboleva A, Tomashevsky M, Marshall KA, et al. NCBI GEO: archive for high-throughput functional genomic data. Nucleic Acids Res. 2009;37:D885–D890. doi: 10.1093/nar/gkn764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pertea G, Huang X, Liang F, Antonescu V, Sultana R, Karamycheva S, Lee Y, White J, Cheung F, Parvizi B, et al. TIGR Gene Indices clustering tools (TGICL): a software system for fast clustering of large EST datasets. Bioinformatics. 2003;19:651–652. doi: 10.1093/bioinformatics/btg034. [DOI] [PubMed] [Google Scholar]
- 13.Youens-Clark K, Buckler E, Casstevens T, Chen C, Declerck G, Derwent P, Dharmawardhana P, Jaiswal P, Kersey P, Karthikeyan A, et al. Gramene database in 2010: updates and extensions. Nucleic Acids Res. 2010:D1085–D1094. doi: 10.1093/nar/gkq1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Swarbreck D, Wilks C, Lamesch P, Berardini TZ, Garcia-Hernandez M, Foerster H, Li D, Meyer T, Muller R, Ploetz L, et al. The Arabidopsis Information Resource (TAIR): gene structure and function annotation. Nucleic Acids Res. 2008;36:D1009–D1014. doi: 10.1093/nar/gkm965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schaeffer ML, Harper LC, Gardiner JM, Andorf CM, Campbell DA, Cannon EKS, Sen TZ, Lawrence CJ. MaizeGDB: curation and outreach go hand-in-hand. Database. 2011;2011 doi: 10.1093/database/bar022. doi:10.1093/database/bar022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Güldener U, Mannhaupt G, Münsterkötter M, Haase D, Oesterheld M, Stümpflen V, Mewes H, Adam G. FGDB: a comprehensive fungal genome resource on the plant pathogen Fusarium graminearum. Nucleic Acids Res. 2006;34:D456–D458. doi: 10.1093/nar/gkj026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tang X, Shen L, Dickerson JA. BarleyExpress: a web-based submission tool for enriched microarray database annotations. Bioinformatics. 2005;21:399–401. doi: 10.1093/bioinformatics/bti002. [DOI] [PubMed] [Google Scholar]
- 18.The Plant Ontology Consortium. The Plant Ontology (TM) Consortium and plant ontologies. Comp. Funct. Genomics. 2002;3:137–142. doi: 10.1002/cfg.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gautier L, Møller M, Friis-Hansen L, Knudsen S. Alternative mapping of probes to genes for Affymetrix chips. BMC Bioinformatics. 2004;5:111. doi: 10.1186/1471-2105-5-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Caldo RA, Nettleton D, Wise RP. Interaction-dependent gene expression in Mla-specified response to barley powdery mildew. Plant Cell. 2004;16:2514–2528. doi: 10.1105/tpc.104.023382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hu P, Meng Y, Wise RP. Functional contribution of chorismate synthase, anthranilate synthase, and chorismate mutase to penetration resistance in barley-powdery mildew interactions. Mol. Plant Microbe Interact. 2009;22:311–320. doi: 10.1094/MPMI-22-3-0311. [DOI] [PubMed] [Google Scholar]
- 22.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]