Abstract
As the volume and complexity of data sets archived at NCBI grow rapidly, so does the need to gather and organize the associated metadata. Although metadata has been collected for some archival databases, previously, there was no centralized approach at NCBI for collecting this information and using it across databases. The BioProject database was recently established to facilitate organization and classification of project data submitted to NCBI, EBI and DDBJ databases. It captures descriptive information about research projects that result in high volume submissions to archival databases, ties together related data across multiple archives and serves as a central portal by which to inform users of data availability. Concomitantly, the BioSample database is being developed to capture descriptive information about the biological samples investigated in projects. BioProject and BioSample records link to corresponding data stored in archival repositories. Submissions are supported by a web-based Submission Portal that guides users through a series of forms for input of rich metadata describing their projects and samples. Together, these databases offer improved ways for users to query, locate, integrate and interpret the masses of data held in NCBI's archival repositories. The BioProject and BioSample databases are available at http://www.ncbi.nlm.nih.gov/bioproject and http://www.ncbi.nlm.nih.gov/biosample, respectively.
INTRODUCTION
The National Center for Biotechnology Information (NCBI) hosts several primary archive databases that store numerous categories of biological data produced by the research community. Data types include, but are not limited to, full and partial genomes, transcriptomes, epigenomes, genetic variation and phenotype data. The NCBI and International Nucleotide Sequence Database Collaboration (INSDC) (1) archival databases hosting these data include GenBank (2), SRA (3), GEO (4), Epigenomics (5), dbSNP (6), dbVar (7) and dbGaP (8).
As the diversity, complexity, inter-relatedness and rate of generation of these data continue to grow, it is becoming increasingly important to organize and annotate the data such that users can more easily locate, understand and analyze data that are relevant to their interests. The BioProject and BioSample databases were recently initiated to help address these needs by facilitating the capture and management of structured metadata and data for diverse biological research projects and samples represented in NCBI's archival databases.
The BioProject database replaces NCBI's Genome Project database and reflects an expansion of project scope, a redesigned database structure and a redesigned website. BioProject serves as a central portal in which to represent the higher order organization, description and classification of data submitted across several NCBI archival databases. It constitutes a flexible framework in which to describe a project's scope and objectives, to group related projects and subprojects and to collate derived data records which would be otherwise dispersed. The BioSample database provides a dedicated area in which to describe the biological materials under investigation in a project and promotes the use of structured and consistent sample attribute descriptions. BioProject and BioSample objects can be reciprocally linked with each other and to corresponding experimental data within any of the archival databases. Neither database is limited by taxonomy and as such includes information spanning eukaryotes, prokaryotes and environmental samples. To support these databases and the primary data archives at NCBI, a new NCBI Submission Portal is being developed to guide and encourage submitters to provide rich project and sample metadata along with their experimental data.
Together, the BioProject and BioSample databases and Submission Portal represent a proactive approach by NCBI to organize and integrate data across interdisciplinary resources and to obtain a rich set of contextual metadata from data producers. These databases allow users to query across many NCBI archives at either the project or sample level to retrieve experimental data sets relevant to their interests. Improved data organization and metadata increase the value of data, promoting more extensive reuse and reanalysis, allowing data to be more easily aggregated and examined from different perspectives, ultimately facilitating novel insights and discoveries across a broad range of biological fields including biomedicine, ecology and evolution.
BIOPROJECT DATABASE OVERVIEW
NCBI's BioProject database represents a higher order organization of biological research projects and availability of the resulting data in several archival databases which are maintained by members of the INSDC. The BioProject database organizes metadata for research projects for which a large volume of data is anticipated and provides a central portal to access the data once it is deposited into an archival database. A BioProject encompasses biological data related to a single initiative, originating from a single organization or from a consortium of coordinating organizations. Records are primarily defined by submitters using the Submission Portal described below but may also be defined by funding source or NCBI staff. A unique BioProject accession number is assigned to each submitted project. Submitters reference this accession when depositing corresponding BioSample records or experimental data into the archival databases. Indeed, a BioProject accession is now a prerequisite for submission of several data types including dbVar structural variation data and whole genome assemblies. BioProject is an INSDC (1) database and, as such, project data are exchanged regularly between the international partners.
The BioProject database provides a dedicated environment in which to:
find distinct data types for a registered project
find projects that are related by a number of different metrics including organism, submitter, data type, collaboration
link project information to experimental data across multiple resources
access information about data availability for a project (sub-project or umbrella collaboration)
support cross-database queries by project identifier
BioProject types, metadata and hierarchies
The resource supports a variety of projects in terms of type and complexity, ranging from a focused genome sequencing project to a large collaboration with multiple data types and subprojects such as genome sequencing, gene expression and epigenetics. The metadata collected includes structured information about the type of research investigation (e.g. genome sequencing, gene expression), project title and goals, the submitting group, environmental sample label or organism and some project data type attributes regarding the sample scope, target and method. This resource also supports registration for a distinct locus_tag prefix, to be used to uniquely identify the genes for genome sequencing projects that will be assembled and annotated.To flexibly describe different types of investigations, information about the sample scope, material used, information captured and methodology are collected (see http://www.ncbi.nlm.nih.gov/books/NBK54364/ for definitions). The combination of these attributes is used to calculate the project data type. For instance, a sequencing project may be from a single individual (monoisolate), multiple individuals of the same species (multi-isolate) or multiple individuals from different species (multispecies). The material being sequenced may be genomic or transcriptomic. The information being captured may be the entire genome or transcriptome, exome or a targeted locus. The method would be sequencing.
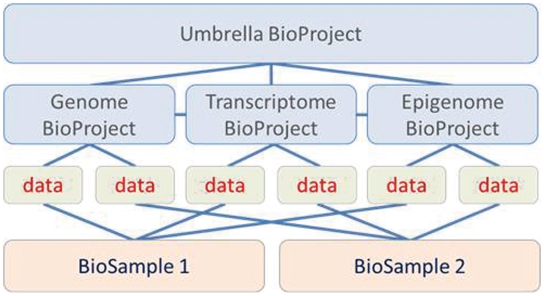
The BioProject framework includes hierarchical linking to represent the complexity inherent in large collaborative research initiatives; in this way, distinct subprojects of a larger initiative (e.g. that apply different methods and yield different types of data) are grouped together by linking to an umbrella project which describes the wider perspective (Figures 1 and 2). This organization supports the ability to collect and organize metadata at both the level of the subproject and the larger collaboration while grouping them together to flexibly support access to the project information and submitted archival data. In addition, projects can be cross-linked to represent other types of relationships between data sets such as reuse of a predefined reference genome standard by an independent research initiative, or biologically relevant relationships or dependencies at the organismal level such as host–parasite. Examples of BioProjects include: (i) PRJNA59457, a metagenome project investigating microbes occurring in a hypersaline lake; (ii) PRJNA20401 and PRJNA13630, organism-specific genome sequencing and transcriptome projects, respectively, for the primate Callithrix jacchus (white-tufted-ear marmoset); or (iii) PRJNA43021, a top level organizational project which groups together subprojects of the NIH Human Microbiome Project. Several other resources are collecting and presenting bio-molecular projects but all of them are limited in scope either by specific organism group [MicrobesOnline (9), IMG (10), GDR (11)] or by project type [GOLD (12)]. BioProject supports a wider scope of projects in terms of type and complexity than any of these resources as it is not limited by organism or data type, i.e. genome, metagenome, expression.
Figure 1.
Schematic depicting how BioProject, BioSample and data objects can be organized and linked. This example is composed of one umbrella project that encompasses three subprojects, each of which generated data derived from two BioSample records. Users can query either the BioProject or the BioSample database to retrieve the relevant records, and then navigate through links to the corresponding experimental data which continue to be stored in NCBI's primary data archives, including GenBank, SRA, dbGaP and GEO. This schematic depicts direct links that can be applied between objects; it does not depict links to corresponding records in other NCBI databases, including PubMed, Gene, Genome and Taxonomy.
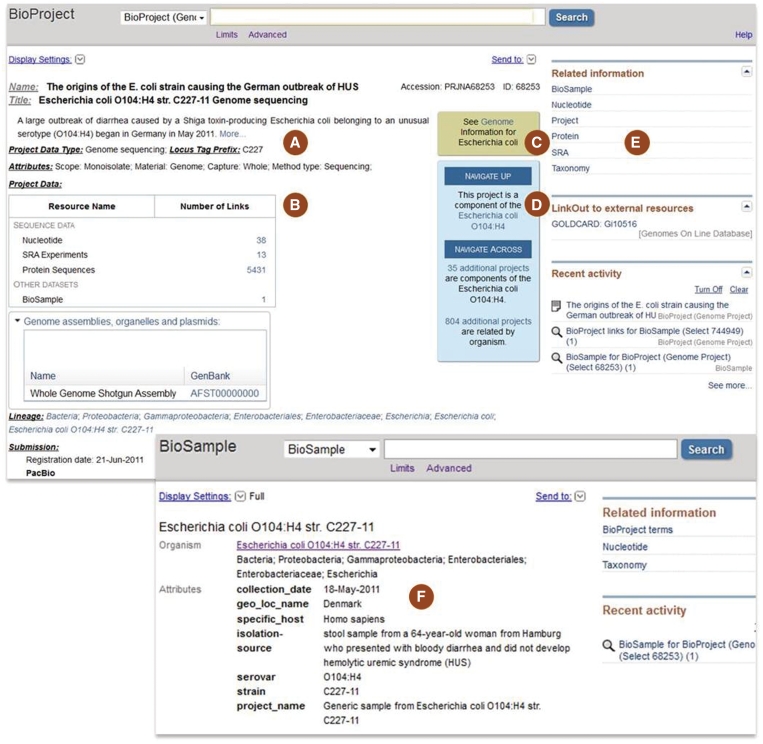
Figure 2.
Screenshot of a Genome Sequencing project that is a component of an umbrella project that encompasses data generated from an E. coli pathogen outbreak (upper panel) (17) and a corresponding sample record (lower panel). The records display the project title, summary, data type, locus_tag prefix and various project attributes including the scope and capture method (A). The Project Data section (B) lists the availability of corresponding sequence and assembly data in the Nucleotide and SRA databases where the data can be downloaded. Navigation panels assist users to link to Genome-level resources for that organism (C), or to ‘Navigate Up’ to the parent umbrella project, or to ‘Navigate across’ to sibling projects that are part of that umbrella project, as well as any additional projects related by organism (D). The ‘Related Information’ panel (E) contains full list of linkages for that record; clicking the BioSample link directs the user to the sample record shown in the lower panel, which lists the attributes that were collected for that sample including the collection date, isolation source, country and strain and serovar (F).
BioProject reports
BioProject full reports present collected project metadata including the project data type (e.g. genome sequencing, transcriptome or gene expression); attributes concerning the sample scope and target, method, and project goals, submitting group, title, organism name or environmental sample label and brief description. If a project is linked to other projects hierarchically, then information about related projects is provided on the page, displayed as links ‘up’ to a higher umbrella project, links ‘down’ from an umbrella to sub-projects or links ‘across’ to sibling projects which share a common umbrella or have other types of relationships. Organism-specific links to the Genome Overview may also be displayed to provide navigation to Entrez Genome. Statistics on data availability are provided based on inclusion of the BioProject identifier on the primary data. If data for a project are available in an archival database and includes the BioProject identifier, then a data table report is included in the BioProject record summarizing those databases with a count of either number of submissions to, or number of records in, the database. For example, SRA reports its content as the number of experiments submitted, whereas the nucleotide database reports its content as the number of accessions. Some projects are registered in the database in advance of data submission and thus do not yet point to any experimental data.
A simple tabular report (http://www.ncbi.nlm.nih.gov/bioproject/browse/) is also available from the home page; this report supports browsing the entire database content as well as filtering to display subsets by project data type, locus_tag prefix or organism. BioProject data are also available for ftp (ftp://ftp.ncbi.nlm.nih.gov/bioproject/) in XML format and a subset of the data is reported as a tab-delimited text file.
Current status
At the time of writing, the BioProject database is populated with almost 12 000 projects derived from more than 3000 organisms. Many of these projects originated from NCBI’s Genome Project resource which was repurposed and redesigned to create this BioProject database. A breakdown of data by project data type is provided in Table 1.
Table 1.
Types of projects in BioProject 5 October 2011
| Project data type | Number |
|---|---|
| Umbrella | 216 |
| Primary submission | 10 712 |
| Assembly | 6 |
| Clone ends | 65 |
| Epigenomics | 9 |
| Exome | 33 |
| Genome sequencing | 9265 |
| Map | 134 |
| Metagenome | 400 |
| Metagenomic assembly | 40 |
| Other | 12 |
| Phenotype or genotype | 3 |
| Proteome | 1 |
| Random survey | 5 |
| Targeted locus (loci) | 90 |
| Transcriptome or gene expression | 588 |
| Variation | 61 |
| RefSeq | 3418 |
As the intra-NCBI database cross-connections are established and enhanced, the number of some project types is expected to increase. For example, the number of ‘Phenotype or genotype’ projects will greatly increase when BioProject is populated with the studies from dbGaP.
BIOSAMPLE DATABASE OVERVIEW
NCBI's BioSample database stores descriptions of the biological materials under examination in a project. Given the huge diversity of sample types handled by NCBI's archival databases, and the fact that appropriate sample descriptions are often dependent on the context of the study, the definition of what a BioSample represents is deliberately flexible. Typical examples of a BioSample include a cell line, a primary tissue biopsy, an individual organism or an environmental isolate. Despite sample heterogeneity, the BioSample database presents new opportunities for improved capture and harmonization of sample descriptions across NCBI databases. It provides a dedicated environment in which to:
capture sample metadata in a structured way by promoting use of controlled vocabularies for sample attribute field names;
link sample information to corresponding experimental data across multiple archival databases;
reduce submitter burden by enabling one-time upload of a sample description, then referencing that sample as appropriate when making data deposits to other archives; and
support cross-database queries by sample description.
Attribute name dictionaries
A major component of a BioSample record is the sample attributes section. Attributes define the material under investigation and can include sample characteristics such as cell type, collection site and phenotypic information like disease state. Since a single sample can be used in multiple experiments and be subjected to different technologies and treatments, information regarding such methodology aspects generally appears on the experimental data records. BioSample attributes are captured as structured name: value pairs, for example, tissue: liver or lat_lon: −39.0, −77.1. The database supports and encourages use of dictionaries of attribute names and synonyms but can also accept any custom attributes that the submitter chooses to provide. While controlled vocabularies are supported for attribute field names, formal ontologies for attribute values are not currently supported. Specific attribute name dictionaries may be utilized for specific sample types. Thus, in the Submission Portal, when a submitter specifies what type of sample they have, the corresponding dictionary is used to drive the request for appropriate attributes. Besides a general dictionary of common core attributes, the first targeted dictionaries implemented in the NCBI BioSample database are the MiXS minimum information checklists for standardizing descriptions of genomes, metagenomes and targeted locus sequences as recently developed by the Genomics Standards Consortium (13). These checklists comprise several packages in which relevant environmental and epidemiological data fields allow for a complete description of the samples being sequenced. The BioSample database is extendible in that dictionary terms and checklists can be added as new standards develop. By guiding and encouraging submitters to use attribute dictionaries in this way, it can be expected that the descriptions for samples deposited via this route will converge and become more consistent over time. This move to broader standardization will in turn assist users to query across NCBI databases for data derived from samples that have specific attributes and values.
BioSample relationships and linking
The BioSample database supports capture of various types of relationships between samples. For example, for samples that represent cell lines derived from individuals with known family relations, pedigree information can be captured and used to group related samples together, facilitating linking to additional relevant records. In addition to these intra-database relationship links, inter-database links are employed to link samples to corresponding experimental data within the primary data archives as well as to the BioProject(s) in which they participate.
Reference BioSample records
While many samples can be considered unique and are used only once, other samples, like commercial cell lines, are re-used over and over again by the research community. We are working with major cell line vendors, including ATCC and Coriell, to generate official representations of commonly used and highly referenced samples. These are Reference BioSamples, so submitters who use these samples may bypass BioSample submission and simply reference relevant Reference BioSample records when depositing experimental data in any of NCBI's primary data archives. Also, efforts will be made to map existing data sets from across NCBI archives to Reference BioSample records. Consequently, these records will serve as hubs from which users can quickly locate a multitude of diverse data sets and projects derived from a given sample. Reference BioSample data will be exchanged and their accession numbers shared with our partners at the European Bioinformatics Institute (EBI) who are establishing a similar BioSample database (14). All Reference BioSample data will be accessible from both the EBI and NCBI databases.
Clinical samples
The BioSample database does not support controlled access mechanisms and thus cannot host human clinical samples that may have associated privacy concerns. Rather, clinical samples will continue to be deposited in NCBI's dbGaP database (8). The dbGaP and BioSample databases are collaborating to present sanitized versions of these data in BioSample, that is, versions where sensitive data attributes are omitted. This allows users to locate these data in BioSample, and then apply to dbGaP for access to the full descriptions as necessary.
Current status
At the time of writing, the BioSample database is populated with over 600 000 original submitter-supplied sample descriptions extracted directly from SRA, sanitized dbGaP samples, GenBank EST and GSS libraries and some GenBank MiXS-compliant samples. As such, while these records may be searched with keywords and attribute names to locate data of interest, it should be noted that these data are generally not curated and were submitted without guidance of the recently implemented BioSample dictionary attributes described above. Going forward, as usage of the NCBI Submission Portal and attribute dictionaries become more widespread, and as Reference BioSamples are loaded, sample descriptions should improve and become more consistent over time.
DATABASE ORGANIZATION AND IMPLEMENTATION
BioProject and BioSample data are stored in separate Microsoft SQL servers and encoded in XML. Databases are structured for efficient storage, tracking and fast retrieval. Figure 1 provides an overview and description of how BioProject, BioSample and experimental data objects may be organized and linked to each other. All BioProject and BioSample metadata are indexed into NCBI's Entrez search and retrieval system.
SUBMISSION PORTAL
The BioProject and BioSample databases provide the infrastructure in which to capture well-annotated, structured metadata that give context to the underlying data, but ultimately it is the researchers who are responsible for providing this information. Thus, it is essential to provide an effective mechanism by which data may be deposited. The NCBI Submission Portal, available at http://submit.ncbi.nlm.nih.gov, is being developed to guide submitters into supplying quality metadata via user-friendly interfaces that balance the need for efficient data input while promoting provision of rich and consistent descriptions. Depositing data through the Submission Portal ensures that submissions are syntactically accurate, promotes capture of metadata using controlled vocabularies and enables linking and aggregation of related objects. However, the BioProject and BioSample databases are submitter-driven primary data archives and submitters are responsible for the quality and content of their deposits. Database staff respond to queries and report errors but, as with other primary data archives, submitted data are not subject to extensive curation.
Use of the Submission Portal requires authentication. Several login options are supported including various National Institutes of Health accounts and a general NCBI PDA (Primary Data Archives) account. Once logged in, the submitter can deposit BioProject and/or BioSample information via the relevant submission wizard, each of which is composed of a series of forms guiding input of descriptive metadata and object relations. Each portal provides a list of the submitter's previously created submissions with some status information and a button to initiate a new submission. A submission may be started, set aside and completed at a later time by signing back into the Submission Portal and selecting the incomplete submission.
In the BioProject portal, submitters describe their initiative and the types of data that they intend to submit. Fields are provided to capture project data type, as well as sample scope, material, capture method and methodology and finally the objectives of the study. Information about the organism or organisms being studied is also collected. One can also input relevant URLs, funding information, consortium information and links and other information. The submitter is asked to provide a description of their project that will be displayed in Entrez BioProject. Once the project is accepted, the submitter receives a BioProject accession and locus_tag prefix, if the project objectives include annotated genome submissions, both of which should be referenced when submitting corresponding experimental data to the archival databases.
In the BioSample portal, submitters describe the biological material under investigation in their project. After specifying the sample type, the user is presented with a list of required and optional attribute fields to fill in, as well as the opportunity to supply any number of custom-descriptive attributes. For example, if a submitter specifies that their sample is a pathogen, they are required to input information about collection locality and date, host, if applicable and isolation source (food, faeces, etc.). In addition, submitters are prompted with additional fields describing the host, disease state, etc. A batch input mechanism using spreadsheets tailored for the sample type is also supported. Authenticated users can provide associations between samples and registered projects, as well as relevant URLs and publications. Once the sample is accepted, the submitter receives a BioSample accession number which should be referenced when submitting corresponding experimental data to the archival databases.
SEARCH AND RETRIEVE DATA
Effective searches may be accomplished using the search box on the BioProject and BioSample home pages at http://www.ncbi.nlm.nih.gov/bioproject and http://www.ncbi.nlm.nih.gov/biosample, respectively. As with other NCBI Entrez databases (15,16), a simple free text keyword search is often sufficient to locate relevant data. However, BioProject and BioSample data are extensively indexed under many fields, meaning that users can refine their search by constructing fielded queries. Some useful search fields are listed in Table 2. Users can write and execute their own search statements directly in the search boxes or, alternatively, use the Advanced search pages which assist users to construct complex multi-part fielded search statements or activate Limits to restrict retrievals to popular search categories. In addition, the BioProject database supports a browse By Project Attributes page that allows users to browse BioProjects in tabular format and to sort and filter by organism, project types and project data type.
Table 2.
Selected search fields and example queries
| Database | Find by … | Example search ‘term[field]’ |
|---|---|---|
| BioProject and BioSample | organism or taxonomic class | insecta[Organism] |
| BioProject | project data type | metagenome[Project Data Type] |
| BioProject | publication | 10473380[Pubmed ID] |
| BioProject | submitter organization or consortium | JGI[Submitter Organization] |
| BioProject | sample scope | scope environment[Properties] |
| BioProject | material used | material transcriptome[Properties] |
| BioProject and BioSample | database identifier | PRJNA33823 or PRJNA33823 [bioproject] or 33823[uid] or 33823[bioproject] |
| BioSample | attribute name | age[Attribute Name] |
| BioSample | attribute and value | cell line GM10847[Attribute] |
| BioSample | data source | source coriell[Properties] |
Multi-part queries may be constructed by specifying the search terms, their fields and the Boolean operations AND, OR, NOT.
BioProject and BioSample records can be accessed by following Entrez links from another NCBI database. As depicted in Figure 1, BioProject and BioSample data are reciprocally linked to corresponding data in the archival databases. This allows users to link to, e.g. corresponding genome assembly records in Entrez Nucleotide or raw sequence reads in SRA where the data can be downloaded (see Figure 2 for location of links). Records are also linked to relevant PubMed, Gene, Genome and Taxonomy records, further facilitating navigation across diverse data domains.
To download BioProject and BioSample descriptions, users can take advantage of the Send to: feature on the search results pages, which allows download of individual or batch BioProject and BioSample retrievals in text or XML formats. Also, BioProject data are available for bulk download in XML, or as a summary tab-delimited text file, from the FTP site at ftp://ftp.ncbi.nlm.nih.gov/bioproject/.
Furthermore, programmatic query, linking and download functions are available using a suite of NCBI programs called Entrez Utilities (http://www.ncbi.nlm.nih.gov/books/NBK25501/).
CONCLUSIONS AND FUTURE DEVELOPMENTS
As the capacity and complexity of biological data sets expands, databases face new challenges in ensuring that the information is adequately organized and described. The NCBI BioProject and BioSample databases are being developed to help address these challenges by providing the means by which data generators can organize and describe a broad range of study designs and sample types, and link to corresponding sets of experimental data in archival databases.
Archival databases must rely heavily on submitters to provide appropriate annotations and context for their projects and samples. The aim of the NCBI Submission Portal is to provide an intuitive interface that balances the requirement for straightforward data input, while encouraging provision of rich and consistent metadata across multiple archival databases and data types. There is a growing appreciation in the community that metadata, in addition to the actual data itself, is essential for interpreting experimental results, for example, when attempting to ascribe function and process to genes, or correlating data with phenotype information. Improved metadata and increased sharing of terminologies across databases provide greater opportunities for data aggregation and making the scientific connections that help transform data into knowledge.
Development of these databases and the Submission Portal is on-going. Many technical refinements are currently being worked on including expanding integration and linking with other databases at NCBI, EBI and DDBJ, improving visualization of object relationships and hierarchies, extending support for controlled vocabularies and checklists for specific BioSample types, enabling batch submission of multi-part projects, building curatorial interfaces, enhancing BioProject web summaries and implementing special reports for funding agencies.
FUNDING
Funding for Open Access charge: Intramural Research Program of the National Institutes of Health, National Library of Medicine.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors thank our EBI colleagues including Alvis Brazma, Helen Parkinson and Ewan Birney for valuable discussions. We also wish to thank the many NCBI staff members who have contributed to discussions and provided data or feedback on these resources, in particular, Mike Feolo, Martin Shumway, Anjanette Johnston, Christopher O'Sullivan, William Klimke, Vasuki Palanigobu, Reza Safarnejad and Anatoly Mnev.
REFERENCES
- 1.Cochrane G, Karsch-Mizrachi I, Nakamura Y. The International Nucleotide Sequence Database Collaboration. Nucleic Acids Res. 2011;39:D15–D18. doi: 10.1093/nar/gkq1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW. GenBank. Nucleic Acids Res. 2011;39:D32–D37. doi: 10.1093/nar/gkq1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shumway M, Cochrane G, Sugawara H. Archiving next generation sequencing data. Nucleic Acids Res. 2010;38:D870–D871. doi: 10.1093/nar/gkp1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barrett T, Troup DB, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH, Sherman PM, et al. NCBI GEO: archive for functional genomics data sets–10 years on. Nucleic Acids Res. 2011;39:D1005–D1010. doi: 10.1093/nar/gkq1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fingerman IM, McDaniel L, Zhang X, Ratzat W, Hassan T, Jiang Z, Cohen RF, Schuler GD. NCBI Epigenomics: a new public resource for exploring epigenomic data sets. Nucleic Acids Res. 2011;39:D908–D912. doi: 10.1093/nar/gkq1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sherry ST, Ward MH, Kholodov M, Baker J, Phan L, Smigielski EM, Sirotkin K. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Church DM, Lappalainen I, Sneddon TP, Hinton J, Maguire M, Lopez J, Garner J, Paschall J, DiCuccio M, Yaschenko E, et al. Public data archives for genomic structural variation. Nat. Genet. 2010;42:813–814. doi: 10.1038/ng1010-813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mailman MD, Feolo M, Jin Y, Kimura M, Tryka K, Bagoutdinov R, Hao L, Kiang A, Paschall J, Phan L, et al. The NCBI dbGaP database of genotypes and phenotypes. Nat. Genet. 2007;39:1181–1186. doi: 10.1038/ng1007-1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dehal PS, Joachimiak MP, Price MN, Bates JT, Baumohl JK, Chivian D, Friedland GD, Huang KH, Keller K, Novichkov PS, et al. MicrobesOnline: an integrated portal for comparative and functional genomics. Nucleic Acids Res. 2010;38:D396–D400. doi: 10.1093/nar/gkp919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Markowitz VM, Szeto E, Palaniappan K, Grechkin Y, Chu K, Chen IM, Dubchak I, Anderson I, Lykidis A, Mavromatis K, et al. The integrated microbial genomes (IMG) system in 2007: data content and analysis tool extensions. Nucleic Acids Res. 2008;36:D528–D533. doi: 10.1093/nar/gkm846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jung S, Jesudurai C, Staton M, Du Z, Ficklin S, Cho I, Abbott A, Tomkins J, Main D. GDR (Genome Database for Rosaceae): integrated web resources for Rosaceae genomics and genetics research. BMC Bioinformatics. 2004;5:130. doi: 10.1186/1471-2105-5-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liolios K, Chen IM, Mavromatis K, Tavernarakis N, Hugenholtz P, Markowitz VM, Kyrpides NC. The Genomes On Line Database (GOLD) in 2009: status of genomic and metagenomic projects and their associated metadata. Nucleic Acids Res. 2010;38:D346–D354. doi: 10.1093/nar/gkp848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yilmaz P, Kottmann R, Field D, Knight R, Cole JR, Amaral-Zettler L, Gilbert JA, Karsch-Mizrachi I, Johnston A, Cochrane G, et al. Minimum information about a marker gene sequence (MIMARKS) and minimum information about any (x) sequence (MIxS) specifications. Nat. Biotechnol. 2011;29:415–420. doi: 10.1038/nbt.1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gostev MFA, Brandizi M, Fernandez-Banet J, Sarkans U, Brazma A, Parkinson H. The BioSample database (BioSD) at the European Bioinformatics Institute. Nucleic Acids Res. 2012;40:D64–D70. doi: 10.1093/nar/gkr937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sayers EW, Barrett T, Benson DA, Bolton E, Bryant SH, Canese K, Chetvernin V, Church DM, DiCuccio M, Federhen S, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2011;39:D38–D51. doi: 10.1093/nar/gkq1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schuler GD, Epstein JA, Ohkawa H, Kans JA. Entrez: molecular biology database and retrieval system. Methods Enzymol. 1996;266:141–162. doi: 10.1016/s0076-6879(96)66012-1. [DOI] [PubMed] [Google Scholar]
- 17.Rasko DA, Webster DR, Sahl JW, Bashir A, Boisen N, Scheutz F, Paxinos EE, Sebra R, Chin CS, Iliopoulos D, et al. Origins of the E. coli strain causing an outbreak of hemolytic-uremic syndrome in Germany. N. Engl. J Med. 2011;365:709–717. doi: 10.1056/NEJMoa1106920. [DOI] [PMC free article] [PubMed] [Google Scholar]