Abstract
Linear motifs are short, evolutionarily plastic components of regulatory proteins and provide low-affinity interaction interfaces. These compact modules play central roles in mediating every aspect of the regulatory functionality of the cell. They are particularly prominent in mediating cell signaling, controlling protein turnover and directing protein localization. Given their importance, our understanding of motifs is surprisingly limited, largely as a result of the difficulty of discovery, both experimentally and computationally. The Eukaryotic Linear Motif (ELM) resource at http://elm.eu.org provides the biological community with a comprehensive database of known experimentally validated motifs, and an exploratory tool to discover putative linear motifs in user-submitted protein sequences. The current update of the ELM database comprises 1800 annotated motif instances representing 170 distinct functional classes, including approximately 500 novel instances and 24 novel classes. Several older motif class entries have been also revisited, improving annotation and adding novel instances. Furthermore, addition of full-text search capabilities, an enhanced interface and simplified batch download has improved the overall accessibility of the ELM data. The motif discovery portion of the ELM resource has added conservation, and structural attributes have been incorporated to aid users to discriminate biologically relevant motifs from stochastically occurring non-functional instances.
INTRODUCTION
Short linear motifs (SLiMs, LMs or MiniMotifs) are regulatory protein modules characterized by their compact interaction interfaces (the affinity and specificity determining residues are usually encoded between 3 and 11 contiguous amino acids (1)) and their enrichment in natively unstructured, or disordered, regions of proteins (2). As a result of limited intermolecular contacts with their interaction partners, SLiMs bind with relatively low affinity (in the low-micromolar range), an advantageous attribute for use as transient, conditional and tunable interactions necessary for many regulatory processes. Due to the limited number of mutations necessary for the genesis of a novel motif, SLiMs are amenable to convergent evolution, functioning as a driver of network evolution by adding novel interaction interfaces, and thereby new functionality, to proteins. This evolutionary plasticity facilitates the rapid proliferation within a proteome, and as a result, motif use is ubiquitous in higher eukaryotes.
SLiMs play an important role for many regulatory processes such as signal transduction, protein trafficking and post-translational modification (3,4). Their importance to the correct functionality of the cell is also reflected by the outcome of motif deregulation. For example, point mutations in SLiMs have been shown to lead severe pathologies such as ‘Noonan-like syndrome’ (5), ‘Liddle’s syndrome’ (6) or ‘Retinitis pigmentosa’ (7). Furthermore, mimicry of linear motifs by viruses to hijack their hosts’ existing cellular machinery plays an important role in many viral life cycles (8). However, despite their obvious importance to eukaryotic cell regulation, our understanding of SLiM biology is relatively limited, and it has been suggested that, to date, we have only discovered a small portion of the human motifs (9).
Several resources are devoted to the annotation and/or detection of SLiMs [Prosite (10), MiniMotifMiner (11) and Scansite (12)]. Here, we report on the 2012 status of the Eukaryotic Linear Motif database.
THE ELM RESOURCE
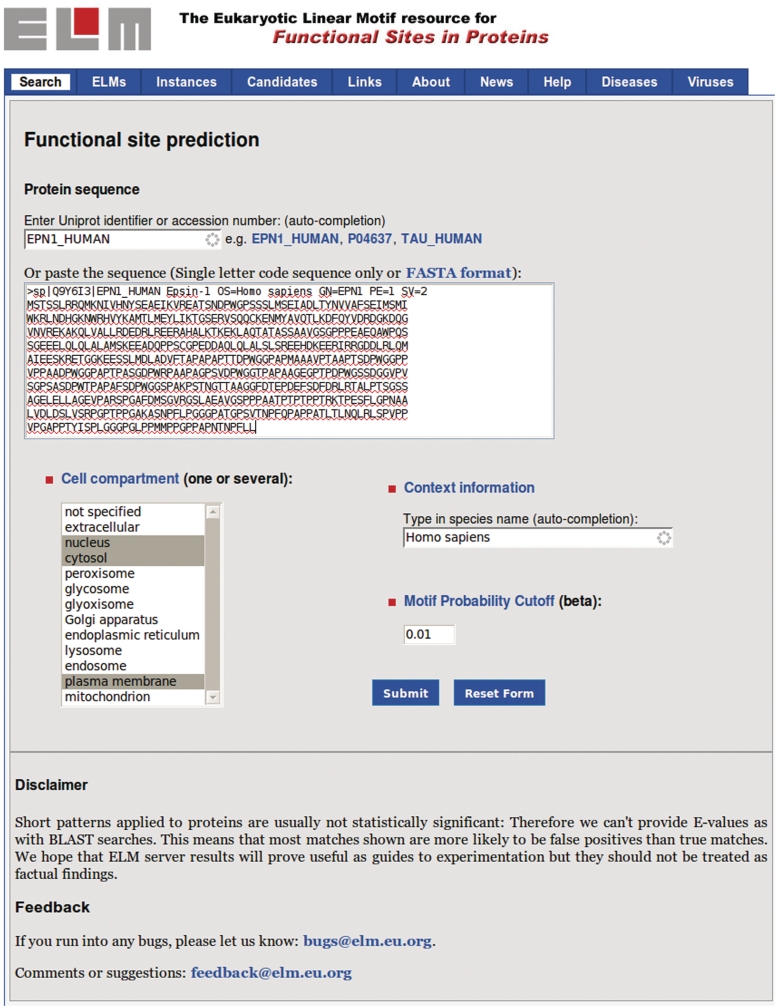
The ELM initiative (http://elm.eu.org) has focused on gathering, storing and providing information about short linear motifs since 2003. It was established as the first manually annotated collection of SLiM classes and as a tool for discovering linear motif instances in proteins (13). As it was mainly focused on the eukaryotic sequences, it was termed the Eukaryotic Linear Motif resource, usually shortened to ELM. The ELM resource consists of two applications: the ELM database of curated motif classes and instances, and the motif detection pipeline to detect putative SLiM instances in query sequences. In the ELM database, SLiMs are annotated as ‘ELM classes’, divided into four ‘types’: cleavage sites (CLV), ligand binding sites (LIG), sites of post-translational modification (MOD) and subcellular targeting sites (TRG) (Table 1). Currently, the ELM database contains 170 linear motif classes with more than 1800 motif instances linked to more than 1500 literature references (Table 1). Each class is described by a regular expression capturing the key specificity and affinity determining amino acid residues. A regular expression is a computer-readable term for sequence annotation and is used by the ELM motif detection pipeline to scan proteins for putative instances of annotated ELM classes. The search form for sequence input is shown in Figure 1, while the results page showing the putative and annotated instances is illustrated in Figure 2.
Table 1.
Summary of data stored in the ELM databasea
| Number of functional site entries | ELM motif classes | ELM motif instances | Links to PDB structures | GO terms | Pubmed links | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Totals | 115 | 170 | 1840 | 195 | 340 | 1561 | ||||
| By category | LIG | 111 | Human | 1004 | ||||||
| MOD | 30 | Mouse | 160 | Biological process | 173 | From ELM motif | 787 | |||
| TRG | 21 | Rat | 102 | |||||||
| CLV | 8 | Fly | 67 | Cell compartment | 74 | From instance | 1071 | |||
| Yeast | 90 | |||||||||
| Other | 417 | Molecular function | 93 | |||||||
aAs of October 2011.
Figure 1.
ELM start page. The user can submit a query sequence to the motif detection pipeline either as UniProt accession number or in FASTA format. Filtering criteria such as taxonomic range or cellular compartment should be activated to limit the resulting list of SLiM instances.
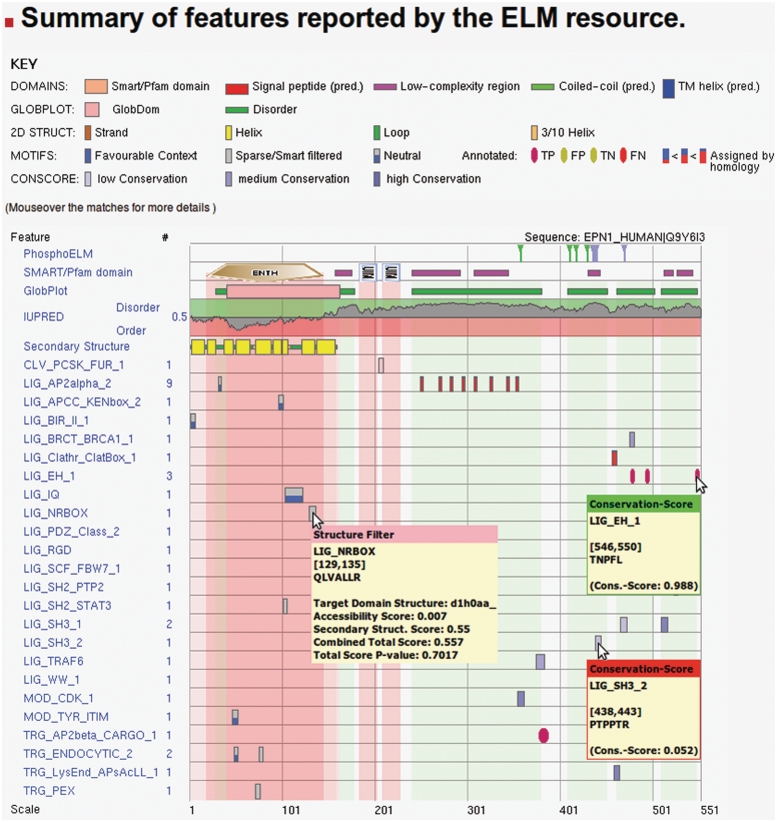
Figure 2.
ELM motif detection pipeline output page. The top legend explains the different colors/symbols used. The graphical output of ELM concentrates the output of multiple sequence classification algorithms; phosphorylation sites from Phospho.ELM, protein domains detected by SMART/Pfam, disorder predictions by GlobPlot and IUPred and secondary structure (18). The lower part contains the annotated and putative ELM instances for the given protein sequence (Epsin1, UniProt accession Q9Y6I3). The background is colored according to the structural information available. Each box represents one ELM instance, the color of which indicates the likelihood that this instance is functional: grey instances are buried within structured regions, while shades of blue represent instances outside of structured regions and hint on sequence conservation, with pale blue representing weak sequence conservation and dark blue indicating strong sequence conservation. Red ellipses or boxes mark instances that are annotated in the query sequence or a homologous sequence, respectively.
The ELM resource is powered by a PostgreSQL relational database for data storage and a PYTHON web framework for data retrieval/visualization. The main tables within the database contain information about ELM classes, ELM instances, sequences, references, taxonomy and links to other databases [the database structure is described in greater detail in (14)].
New ELM classes
Since the last release (14), 24 new ELM classes have been added to the ELM database (Table 1) and several more have been updated. One of the newly annotated motif classes is the AGC kinase docking motif (LIG_AGCK_PIF), consisting of three distinct classes. It is present in the non-catalytic C-terminal tail of AGC kinases that constitute a family of serine/threonine kinases consisting of 60 members that regulate critical processes, including cell growth and survival. Deregulation of these enzymes is a causative factor in different diseases such as cancer and diabetes. The motif interacts with the PDK1 Interacting Fragment (PIF) pocket in the kinase domain of AGC kinases. It mediates intramolecular binding to the PIF pocket, serving as a cis-activating module together with other regulatory sequences in the C-tail. Interestingly, in some kinases the motif also acts as a PDK1 docking site that trans-activates PDK1, which itself lacks the regulatory C-tail, by interacting with the PDK1 PIF pocket. PDK1 in turn will phosphorylate and activate the docked kinase. Other novel classes (Table 2) include phosphodegrons, which are important mediators of phosphorylation-dependent protein destruction, and the LYPxL motif, which is involved in endosomal sorting of membrane proteins but is also implicated in retrovirus budding.
Table 2.
List of novel ELM classesa
| Identifier | Description |
|---|---|
| LIG_Actin_WH2_1 | Motifs, present in proteins in several repeats, which mediate binding to the hydrophobic cleft created by subdomains 1 and 3 of G-actin |
| LIG_Actin_WH2_2 | |
| LIG_Actin_RPEL_3 | |
| LIG_AGCK_PIF_1 | The AGCK docking motif mediates intramolecular interactions to the PDK1 Interacting Fragment (PIF) pocket, serving as a cis-activating module |
| LIG_AGCK_PIF_2 | |
| LIG_AGCK_PIF_3 | |
| LIG_BIR_II_1 | IAP-binding motifs are found in pro-apoptotic proteins and function in the abrogation of caspase inhibition by inhibitor of apoptosis proteins in apoptotic cells |
| LIG_BIR_III_1 | |
| LIG_BIR_III_2 | |
| LIG_BIR_III_3 | |
| LIG_BIR_III_4 | |
| LIG_eIF4E_1 | Motif binding to the dorsal surface of eIF4E |
| LIG_eIF4E_2 | |
| LIG_EVH1_3 | A proline-rich motif binding to EVH1/WH1 domains of WASP and N-WASP proteins |
| LIG_HCF-1_HBM_1 | The DHxY Host Cell Factor-1 binding motif interacts with the N-terminal kelch propeller domain of the cell cycle regulator HCF-1 |
| LIG_Integrin_isoDGR_1 | Present in proteins of extracellular matrix which upon deamidation forms biologically active isoDGR motif which binds to various members of integrin family |
| LIG_LYPXL_L_2 | The LYPxL motif binds the V-domain of Alix, a protein involved in endosomal sorting |
| LIG_LYPXL_S_1 | |
| LIG_PAM2_1 | Peptide ligand motif that directly interacts with the MLLE/PABC domain found in poly(A) binding proteins and HYD E3 ubiquitin ligases |
| LIG_PIKK_1 | Motif located in the C terminus of Nbs1 and its homologous interacting with PIKK family members |
| LIG_Rb_pABgroove_1 | The LxxLFD motif binds in a deep groove between pocket A and pocket B of the Retinoblastoma protein |
| LIG_SCF_FBW7_1 | The TPxxS phospho-dependent degron binds the FBW7 F box proteins of the SCF (Skp1-Cullin-Fbox) complex |
| LIG_SCF_FBW7_2 | |
| LIG_SPAK-OSR1_1 | SPAK/OSR1 kinase binding motif acts as a docking site which aids the interaction with their binding partners including the upstream activators and the phosphorylated substrates |
aAs of October 2011.
New ELM instances
Annotated ELM instances serve as representative examples of the respective ELM class. They are also invaluable for the computational analysis and classification of motifs (15). Therefore, special emphasis has been put on the curation of more than 500 novel ELM instances (in 40 different classes) by scanning and annotating more than 400 articles. The number of protein databank (PDB) entries annotated have been increased to 195 (Table 1), meaning that for ∼10% of all instances there is a 3D protein structure annotated, giving more detailed information about the biological context of the respective motif.
NEW FEATURES
The ELM website at http://elm.eu.org can be used in two ways: first, as a front-end to explore the ELM database of curated ELM classes and instances, and second, to run the motif detection pipeline to detect putative SLiM instances in query sequences. Both interfaces have been improved with the most notable changes listed below.
User interface
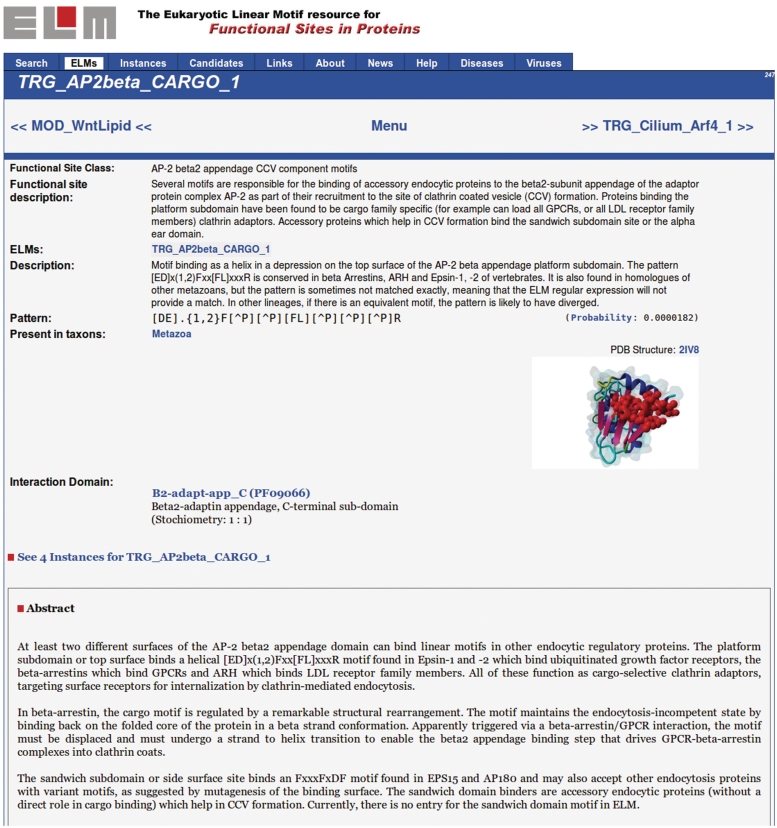
The database user interface, having been stable for many years, has been overhauled and replaced by a novel interface introducing several new features (Figure 1). Up-to-date web technologies have been used to improve the general user experience: the PYTHON framework DJANGO (http://www.djangoproject.com) dynamically creates and serves all HTML pages, while JavaScript was used to make the whole site more interactive and thus improve the user experience. In particular, the ELM detail pages (Figure 3), which hold the most important information about each ELM class including references, regular expression, taxonomic distribution and gene ontology terms (Table 3), have been updated by annotating the protein domain interacting with the respective motif. Where available, a 3D model of representative protein databank structures of linear motif interactions was added to the ELM detail page (Figure 3, top right).
Figure 3.
ELM detail page showing information about the ELM class TRG_AP2beta_CARGO_1.
Table 3.
Main cellular compartments used in ELM annotation
| Count | GO Id | GO term |
|---|---|---|
| 98 | GO:0005829 | Cytosol |
| 69 | GO:0005634 | Nucleus |
| 17 | GO:0005576 | Extracellular |
| 12 | GO:0005794 | Golgi apparatus |
| 10 | GO:0005886 | Plasma membrane |
| 9 | GO:0009898 | Internal side of plasma membrane |
| 9 | GO:0005783 | Endoplasmic reticulum |
| 6 | GO:0005739 | Mitochondrion |
| 5 | GO:0005643 | Nuclear pore |
| 5 | GO:0045334 | Clathrin-coated endocytic vesicle |
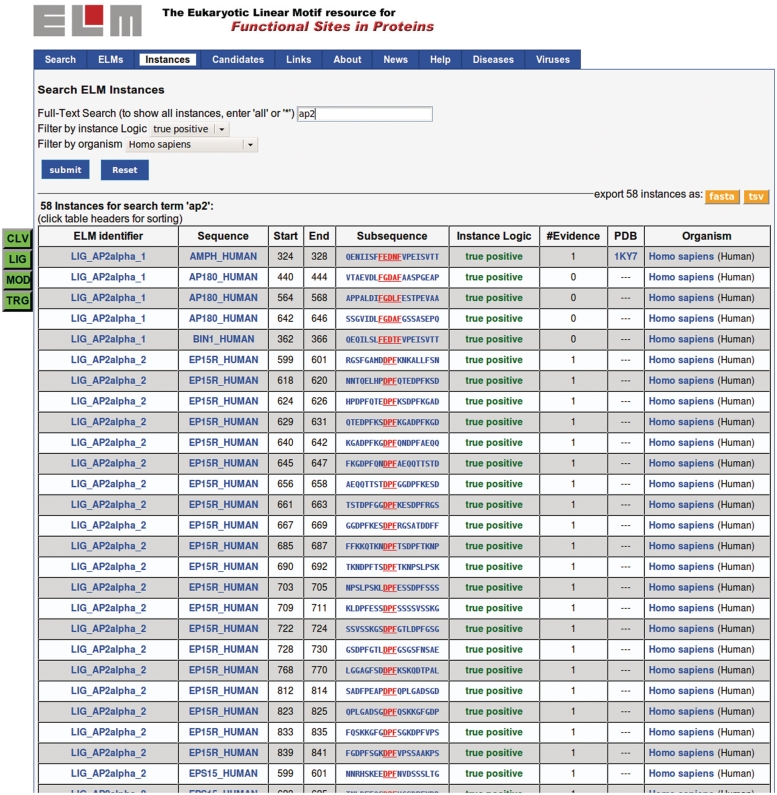
To cope with the increasing amount of annotated classes as well as instances, a novel query interface was introduced to assist the user in finding information of interest. The ELM browser (Figure 4) now features a search interface for free text search. In addition, the search results can also be filtered and reordered using buttons (Figure 4, left side) and table headers, respectively, and be downloaded as tab-separated values (TSV).
Figure 4.
ELM instances browse page. A full-text search (here, search term used was ‘AP2’, filtering for ‘true positive’ instances in taxon ‘Homo sapiens’, yielding 58 instances) assists in finding annotated instances. A search can be restricted to a particular taxonomy or instance logic (top) or ELM class type (buttons on the left). The list can also be exported to TSV or FASTA format for further processing.
Further, improvements to the ELM database include revising the experimental methods used for annotation by using a standardized methods vocabulary [in sync with PSI-MI ontology (16,17)].
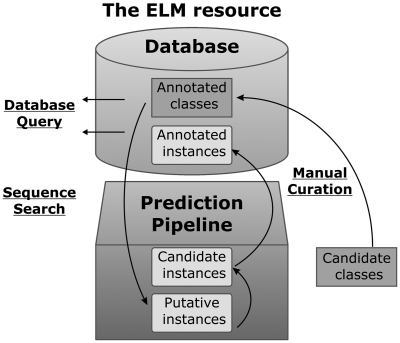
A candidate page has been introduced to display novel ELM classes that have not yet been annotated in detail or are currently undergoing annotation. We invite researchers to send us their feedback and expert opinion on these classes and to contribute novel motif classes that will be added to the candidate page and ultimately be turned into full ELM classes (Figure 5). Minimum requirements are at least one literature reference as well as a short description. In addition, a draft regular expression or a 3D structure showing the relevant interaction would also be helpful. Currently, the number of possible ELM classes on this candidate list (awaiting further annotation) exceeds the number of completely annotated classes, indicating the great demand for further annotation.
Figure 5.
Schema of the ELM resource and data life cycle. Annotated ELM classes, and instances thereof, can be searched by database query. Via sequence search by the motif detection pipeline, annotated ELM classes yield putative instances in query sequences. By adding experimental evidence and references, these putative instances become candidate instances for annotation, and, with further curation, ultimately become fully annotated instances.
Graphical representation of sequence search
The ELM motif detection pipeline scans protein sequences for matches to the regular expressions of annotated ELM classes (Figure 2). The query output combines these putative instances with information from the database (annotated ELM instances) as well as predictions from different algorithms/filters. The ELM resource employs a structural filter (18) to highlight and mask secondary structure elements, as well as SMART (19) to detect protein domains. Furthermore, an additional disorder prediction algorithm (IUPred) (20) has been included to predict ordered/disordered regions within the protein. IUPred uses a cutoff of 0.5 to classify a sequence region as either structured or disordered, with values above this threshold corresponding to disorder, highlighted in green background and lower values indicating structured regions, displayed in red background in the output graph. Disorder and domain information is combined by background coloring to highlight structured regions within the protein, which allows inspection of SLiMs that reside at domain boundaries and emphasizes motifs in disordered regions.
The conservation of linear motifs can help in assessing the functional relevance of putative instances, with functional instances showing higher overall sequence conservation than non-functional ones (21). Therefore, sequence conservation of the query protein is calculated using a tree-based conservation scoring method (22) and highlighted in the graphical output. Here, lighter shades of blue represent low conservation while dark blue shading corresponds to high-sequence conservation. The actual conservation score can be inspected by moving the mouse over the respective ELM instance (Figure 2).
The functionality of linear motifs can be modulated by modifications such as phosphorylation (23,24). To enable the user to investigate phosphorylation data in the context of putative linear motif instances, phosphorylation annotations from the Phospho.ELM resource (25) have been added to the graphical output (Figure 2, top row). The phosphorylated residues are highlighted in different colors (serine: green, threonine: blue, tyrosine: red); each phosphorylation site is linked to a page showing detailed information about the respective modification site from the manually curated data set of the Phospho.ELM resource.
VIRAL INSTANCES
The importance of the short linear motifs in virus–host interactions makes the ELM resource an important tool for the viral research community. For example, Cruz et al. (26) analyzed a protein phosphatase 1 (PP1) docking motif in ‘protein 7’ of transmissible gastroenteritis virus using the ELM class LIG_PP1. This conserved sequence motif mediates binding to the PP1 catalytic subunit, a key regulator of the cellular antiviral defense mechanisms, and is also found in other viral proteomes, suggesting that it might be a recurring strategy to counteract the hosts’ defense against RNA viruses by dephosphorylating eukaryotic translation initiation factor 2α and ultimately ribonuclease L.
To reflect our increasing awareness of viral motifs (8), special focus has been attributed to the annotation of viral instances in the ELM database: in the latest release, more than 200 novel ELM instances found in 84 different viral taxons have been added. The notion of viruses abusing existing SLiMs in their hosts is demonstrated by viral instances being annotated alongside instances in their hosts’ proteins. For example, the ELM class LIG_PDZ_Class_1 contains 12 instances in human proteins but has recently been expanded with 5 instances from 5 different human pathogenic virus proteins.
LINEAR MOTIFS AND DISEASES
The importance of SLiMs is further corroborated by the occurrence of pathologies that are caused by mutations that either mutate existing linear motifs or create novel linear motifs (of undesired function) (27). Examples include ‘Usher's syndrome’ (28), ‘Liddle's Syndrome’ (6) or ‘Golabi-Ito-Hall Syndrome’ (29). The developmental disorder ‘Noonan Syndrome’ can be caused by mutations in Raf-1 that abrogate the interaction with 14-3-3 proteins mediated by corresponding SLiMs and thereby deregulate the Raf-1 kinase activity (30) (the Raf-1 protein sequence features two LIG_14-3-3_1 binding sites that are annotated at 256-261 and 618-623 in the ELM resource). A related disease, ‘Noonan-like Syndrome’, is caused by an S to G mutation at position 2 of the SHOC2 protein, creating a novel myristoylation site (annotated as ELM class MOD_NMyristoyl). This irreversible modification results in aberrant targeting of SHOC2 to the plasma membrane and impaired translocation to the nucleus upon growth factor stimulation (5). More information about the implication of short linear motifs on diseases is collected at http://elm.eu.org/infos/diseases.html.
APPLICATION OF THE ELM RESOURCE
By providing a high-quality, manually curated data set of linear motif classes with experimentally validated SLiM instances, the ELM database has proven to be invaluable to the community: small-scale (single protein) analyzes benefit from the detailed annotation of each ELM class in attributing novel features to proteins of interest. By using in vitro and in vivo studies, von Nandelstadh et al. (31) could validate a PDZ class III motif, detected by ELM at the carboxy terminus of myotilin and the FATZ (calsarcin/myozenin) families. This evolutionarily conserved carboxy-terminal motif mediates binding to PDZ domains of ZASP/Cypher and other Enigma family members (ALP, CLP-36 and RIL) and disruption of these interactions results in myofibrillar myopathies (32). Additionally, ELM annotations can contribute to high-throughput screenings (33) as well as development of novel algorithms (34–36), methods (37) and databases (38). Furthermore, the highly curated data of the ELM resource are used as a benchmarking data set to evaluate the accuracy of prediction algorithms (21,39,40).
For any such analysis, the user should be aware that many matches to ELM regular expressions are false positives. Before conducting experiments based on ELM results, it is strongly advisable to check if a motif match is conserved, exposed in a cell compartment in which the motif is known to be functional. The ELM resource applies several filters to provide the user with such information that should ideally also be supported by the experimental evidence.
SUMMARY
The importance of SLiMs is highlighted by the growing number of instances with relevance to diseases or viruses. Yet, despite their importance and abundance, our understanding of linear motifs is still limited. This is mainly owing to the fact that they are still quite difficult to predict computationally and to investigate experimentally (3,41,42). By better understanding the biology of linear motifs, we hope to increase our insight into diseases and viruses (and vice versa). The ELM resource tries to aid the researcher in the search for putative SLiM instances by providing a feature-rich toolset for sequence analysis. Consequently, with the aforementioned additions and changes, we hope that the ELM resource continues to be a valuable asset to the community.
FUNDING
EMBL international PhD program (to R.J.W.); EMBL Interdisciplinary PostDoc fellowship (EIPOD to N.E.D.); NGFN framework by the Federal Government Department of Education and Science [FKZ01GS0862 (DiGtoP) to M.S. and M.H.]; European Community's Seventh Framework Programme FP7/2009 (SysCilia) (241955 to G.T.) and (SyBoSS) (242129 to K.V.R.); Polish Ministry of Science and Higher Education within Iuventus Plus project (IP2010-0483-70 to M.D.); Biotechnology and Biological Sciences Research Council (BB/F010486/1 to A.C.); Région Alsace and Collège Doctoral Européen (to K.L.); Science Foundation Ireland (08/IN.1/B1864 to G.G.); BBSRC New Investigator Award (BB/I006230/1 to R.J.E.); German Research Foundation (SFB796 Project A2 to H.M.); grants from the Swiss National Science Foundation (to M.O.S.). Funding for open access charge: EMBL.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors would like to thank the users of the ELM resource as well as all colleagues, contributors and annotators of the ELM resource.
REFERENCES
- 1.Davey NE, Van Roey K, Weatheritt RJ, Toedt G, Uyar B, Altenberg B, Budd A, Diella F, Dinkel H, Gibson TJ. Attributes of short linear motifs. Mol Biosyst. 2011 doi: 10.1039/c1mb05231d. September 12 (doi:10.1039/c1mb05231d; epub ahead of print) [DOI] [PubMed] [Google Scholar]
- 2.Fuxreiter M, Tompa P, Simon I. Local structural disorder imparts plasticity on linear motifs. Bioinformatics. 2007;8:950–956. doi: 10.1093/bioinformatics/btm035. [DOI] [PubMed] [Google Scholar]
- 3.Diella F, Haslam N, Chica C, Budd A, Michael S, Brown NP, Trave G, Gibson TJ. Understanding eukaryotic linear motifs and their role in cell signaling and regulation. Front. Biosci. 2008:6580–6603. doi: 10.2741/3175. [DOI] [PubMed] [Google Scholar]
- 4.Gibson TJ. Cell regulation: determined to signal discrete cooperation. Trends Biochem. Sci. 2009;34:471–482. doi: 10.1016/j.tibs.2009.06.007. [DOI] [PubMed] [Google Scholar]
- 5.Cordeddu V, Di Schiavi E, Pennacchio LA, Ma'ayan A, Sarkozy A, Fodale V, Cecchetti S, Cardinale A, Martin J, Schackwitz W, et al. Mutation of SHOC2 promotes aberrant protein N-myristoylation and causes Noonan-like syndrome with loose anagen hair. Nat. Genet. 2009;9:1022–1026. doi: 10.1038/ng.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Furuhashi M, Kitamura K, Adachi M, Miyoshi T, Wakida N, Ura N, Shikano Y, Shinshi Y, Sakamoto K, Hayashi M, et al. Liddle's syndrome caused by a novel mutation in the proline-rich PY motif of the epithelial sodium channel beta-subunit. J. Clin. Endocrinol. Metab. 2005;1:340–344. doi: 10.1210/jc.2004-1027. [DOI] [PubMed] [Google Scholar]
- 7.Deretic D, Schmerl S, Hargrave PA, Arendt A, McDowell JH. Regulation of sorting and post-Golgi trafficking of rhodopsin by its C-terminal sequence QVS(A)PA. Proc. Natl Acad. Sci. USA. 1998;18:10620–10625. doi: 10.1073/pnas.95.18.10620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davey NE, Trave G, Gibson TJ. How viruses hijack cell regulation. Trends Biochem. Sci. 2011;3:159–169. doi: 10.1016/j.tibs.2010.10.002. [DOI] [PubMed] [Google Scholar]
- 9.Neduva V, Linding R, Su-Angrand I, Stark A, de Masi F, Gibson TJ, Lewis J, Serrano L, Russell RB. Systematic discovery of new recognition peptides mediating protein interaction networks. PLoS Biol. 2005;12:e405. doi: 10.1371/journal.pbio.0030405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hulo N, Bairoch A, Bulliard V, Cerutti L, Cuche BA, de Castro E, Lachaize C, Langendijk-Genevaux PS, Sigrist CJ. The 20 years of PROSITE. Nucleic Acids Res. 2008;36:D245–D249. doi: 10.1093/nar/gkm977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rajasekaran S, Balla S, Gradie P, Gryk MR, Kadaveru K, Kundeti V, Maciejewski MW, Mi T, Rubino N, Vyas J, et al. Minimotif miner 2nd release: a database and web system for motif search. Nucleic Acids Res. 2009;37:D185–D190. doi: 10.1093/nar/gkn865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Obenauer JC, Cantley LC, Yaffe MB. Scansite 2.0: proteome-wide prediction of cell signaling interactions using short sequence motifs. Nucleic Acids Res. 2003;13:3635–3641. doi: 10.1093/nar/gkg584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Puntervoll P, Linding R, Gemund C, Chabanis-Davidson S, Mattingsdal M, Cameron S, Martin DM, Ausiello G, Brannetti B, Costantini A, et al. ELM server: a new resource for investigating short functional sites in modular eukaryotic proteins. Nucleic Acids Res. 2003;13:3625–3630. doi: 10.1093/nar/gkg545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gould CM, Diella F, Via A, Puntervoll P, Gemund C, Chabanis-Davidson S, Michael S, Sayadi A, Bryne JC, et al. ELM: the status of the 2010 eukaryotic linear motif resource. Nucleic Acids Res. 2010;38:D167–D180. doi: 10.1093/nar/gkp1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Davey NE, Edwards RJ, Shields DC. Computational identification and analysis of protein short linear motifs. Front. Biosci. 2010;15:801–825. doi: 10.2741/3647. [DOI] [PubMed] [Google Scholar]
- 16.Hermjakob H, Montecchi-Palazzi L, Bader G, Wojcik J, Salwinski L, Ceol A, Moore S, Orchard S, Sarkans U, von Mering C, et al. The HUPO PSI's molecular interaction format–a community standard for the representation of protein interaction data. Nat. Biotechnol. 2004;2:177–183. doi: 10.1038/nbt926. [DOI] [PubMed] [Google Scholar]
- 17.Cote RG, Jones P, Apweiler R, Hermjakob H. The Ontology Lookup Service, a lightweight cross-platform tool for controlled vocabulary queries. BMC Bioinformatics. 2006;7 doi: 10.1186/1471-2105-7-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Via A, Gould CM, Gemund C, Gibson TJ, Helmer-Citterich M. A structure filter for the Eukaryotic Linear Motif Resource. BMC Bioinformatics. 2009;10 doi: 10.1186/1471-2105-10-351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Letunic I, Doerks T, Bork P. SMART 6: recent updates and new developments. Nucleic Acids Res. 2009;37:D229–D232. doi: 10.1093/nar/gkn808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dosztanyi Z, Csizmok V, Tompa P, Simon I. IUPred: web server for the prediction of intrinsically unstructured regions of proteins based on estimated energy content. Bioinformatics. 2005;16:3433–3434. doi: 10.1093/bioinformatics/bti541. [DOI] [PubMed] [Google Scholar]
- 21.Dinkel H, Sticht H. A computational strategy for the prediction of functional linear peptide motifs in proteins. Bioinformatics. 2007;24:3297–3303. doi: 10.1093/bioinformatics/btm524. [DOI] [PubMed] [Google Scholar]
- 22.Chica C, Labarga A, Gould CM, Lopez R, Gibson TJ. A tree-based conservation scoring method for short linear motifs in multiple alignments of protein sequences. BMC Bioinformatics. 2008;9 doi: 10.1186/1471-2105-9-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Balagopalan L, Coussens NP, Sherman E, Samelson LE, Sommers CL. The LAT story: a tale of cooperativity, coordination, and choreography. Cold Spring Harb. Perspect. Biol. 2010;8:a005512. doi: 10.1101/cshperspect.a005512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pawson T, Scott JD. Protein phosphorylation in signaling–50 years and counting. Trends Biochem. Sci. 2005;6:286–290. doi: 10.1016/j.tibs.2005.04.013. [DOI] [PubMed] [Google Scholar]
- 25.Dinkel H, Chica C, Via A, Gould CM, Jensen LJ, Gibson TJ, Diella F. Phospho.ELM: a database of phosphorylation sites–update 2011. Nucleic Acids Res. 2011;39:D261–D267. doi: 10.1093/nar/gkq1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cruz JL, Sola I, Becares M, Alberca B, Plana J, Enjuanes L, Zuniga S. Coronavirus gene 7 counteracts host defenses and modulates virus virulence. PLoS Pathog. 2011;6:e1002090. doi: 10.1371/journal.ppat.1002090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kadaveru K, Vyas J, Schiller MR. Viral infection and human disease–insights from minimotifs. Front. Biosci. 2008;13:6455–6471. doi: 10.2741/3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weil D, El-Amraoui A, Masmoudi S, Mustapha M, Kikkawa Y, Laine S, Delmaghani S, Adato A, Nadifi S, Zina ZB, et al. Usher syndrome type I G (USH1G) is caused by mutations in the gene encoding SANS, a protein that associates with the USH1C protein, harmonin. Hum. Mol. Genet. 2003;5:463–471. doi: 10.1093/hmg/ddg051. [DOI] [PubMed] [Google Scholar]
- 29.Tapia VE, Nicolaescu E, McDonald CB, Musi V, Oka T, Inayoshi Y, Satteson AC, Mazack V, Humbert J, Gaffney CJ, et al. Y65C missense mutation in the WW domain of the Golabi-Ito-Hall syndrome protein PQBP1 affects its binding activity and deregulates pre-mRNA splicing. J. Biol. Chem. 2010;25:19391–19401. doi: 10.1074/jbc.M109.084525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pandit B, Sarkozy A, Pennacchio LA, Carta C, Oishi K, Martinelli S, Pogna EA, Schackwitz W, Ustaszewska A, Landstrom A, et al. Gain-of-function RAF1 mutations cause Noonan and LEOPARD syndromes with hypertrophic cardiomyopathy. Nat. Genet. 2007;8:1007–1012. doi: 10.1038/ng2073. [DOI] [PubMed] [Google Scholar]
- 31.von Nandelstadh P, Ismail M, Gardin C, Suila H, Zara I, Belgrano A, Valle G, Carpen O, Faulkner G. A class III PDZ binding motif in the myotilin and FATZ families binds enigma family proteins: a common link for Z-disc myopathies. Mol. Cell Biol. 2009;3:822–834. doi: 10.1128/MCB.01454-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Selcen D, Engel AG. Mutations in myotilin cause myofibrillar myopathy. Neurology. 2004;8:1363–1371. doi: 10.1212/01.wnl.0000123576.74801.75. [DOI] [PubMed] [Google Scholar]
- 33.Gfeller D, Butty F, Wierzbicka M, Verschueren E, Vanhee P, Huang H, Ernst A, Dar N, Stagljar I, Serrano L, et al. The multiple-specificity landscape of modular peptide recognition domains. Mol. Syst. Biol. 2011;7 doi: 10.1038/msb.2011.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bauer DC, Willadsen K, Buske FA, Le Cao KA, Bailey TL, Dellaire G, Boden M. Sorting the nuclear proteome. Bioinformatics. 2011;13:i7–i14. doi: 10.1093/bioinformatics/btr217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Walsh I, Martin AJ, Di Domenico T, Vullo A, Pollastri G, Tosatto SC. CSpritz: accurate prediction of protein disorder segments with annotation for homology, secondary structure and linear motifs. Nucleic Acids Res. 2011;39:W190–W196. doi: 10.1093/nar/gkr411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lieber DS, Elemento O, Tavazoie S. Large-scale discovery and characterization of protein regulatory motifs in eukaryotes. PLoS One. 2010;12:e14444. doi: 10.1371/journal.pone.0014444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pless O, Kowenz-Leutz E, Dittmar G, Leutz A. A differential proteome screening system for post-translational modification-dependent transcription factor interactions. Nat. Protoc. 2011;3:359–364. doi: 10.1038/nprot.2011.303. [DOI] [PubMed] [Google Scholar]
- 38.Goel R, Muthusamy B, Pandey A, Prasad TS. Human protein reference database and human proteinpedia as discovery resources for molecular biotechnology. Mol. Biotechnol. 2011;1:87–95. doi: 10.1007/s12033-010-9336-8. [DOI] [PubMed] [Google Scholar]
- 39.Edwards RJ, Davey NE, Shields DC. SLiMFinder: a probabilistic method for identifying over-represented, convergently evolved, short linear motifs in proteins. PLoS One. 2007;10:e967. doi: 10.1371/journal.pone.0000967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Edwards RJ, Davey NE, Shields DC. CompariMotif: quick and easy comparisons of sequence motifs. Bioinformatics. 2008;10:1307–1309. doi: 10.1093/bioinformatics/btn105. [DOI] [PubMed] [Google Scholar]
- 41.Perkins JR, Diboun I, Dessailly BH, Lees JG, Orengo C. Transient protein-protein interactions: structural, functional, and network properties. Structure. 2010;10:1233–1243. doi: 10.1016/j.str.2010.08.007. [DOI] [PubMed] [Google Scholar]
- 42.Edwards RJ, Davey NE, Brien KO, Shields DC. Interactome-wide prediction of short, disordered protein interaction motifs in humans. Mol. Biosyst. 2011 doi: 10.1039/c1mb05212h. August 30 (doi:10.1039/c1mb05212h; epub ahead of print) [DOI] [PubMed] [Google Scholar]