Abstract
IntAct is an open-source, open data molecular interaction database populated by data either curated from the literature or from direct data depositions. Two levels of curation are now available within the database, with both IMEx-level annotation and less detailed MIMIx-compatible entries currently supported. As from September 2011, IntAct contains approximately 275 000 curated binary interaction evidences from over 5000 publications. The IntAct website has been improved to enhance the search process and in particular the graphical display of the results. New data download formats are also available, which will facilitate the inclusion of IntAct's data in the Semantic Web. IntAct is an active contributor to the IMEx consortium (http://www.imexconsortium.org). IntAct source code and data are freely available at http://www.ebi.ac.uk/intact.
INTRODUCTION
Understanding the interactions a protein makes with the molecules in its immediate environment, is critical for a full understanding of the processes in which that protein is involved and the mechanisms by which it is regulated. Interaction data can be generated using many different techniques, all of which have their strengths and weaknesses. Many of these techniques can be used in high throughput mode, potentially giving information on several thousand pairs of interacting molecules or identifying over a hundred prey proteins, which may bind to a single bait molecule. To bring together a true picture of the interactions occurring within any living organism, all this data needs to be gathered into central repositories. In a database, each interaction can either be reinforced by additional interaction evidences using other experimental procedures, or identified as an isolated example of this interaction, and as such, potentially false positive data. The IntAct molecular interaction database (http://www.ebi.ac.uk/intact) exists to collect and collate such data. The database undertakes both archival curation of the literature and also actively encourages data producers to deposit interaction data as part of the publication process. The database is compliant with HUPO-PSI data standards and releases data in both the PSI-MI XML 2.5 and PSIMITAB formats (1), either via the website, PSICQUIC web service or from ftp://ftp.ebi.ac.uk/pub/databases/intact/current. All data is made freely available, under the Creative Commons Attribution license. IntAct is implemented using the Java language using a number of external and internal open source libraries. All the software produced by the IntAct developers is free and open source, and can be used, modified and redistributed under the terms of the Apache Software License. This includes the database schema itself. Users are encouraged to join a public mailing list which has been created to support its users and discuss development issues (http://groups.google.com/group/intact-developers).
INTACT CURATION
Curation policy and data types
The information within the IntAct database primarily consists of protein–protein interaction (PPI) data. IntAct is an active member of the IMEx consortium (S. Orchard et al., manuscript in preparation), and the majority of the PPI data within the database is annotated to IMEx standards, as agreed by the IMEx consortium. All such records contain a full description of the experimental conditions in which the interaction was observed. This includes full details of the constructs used in each experiment, such as the presence and position of tags, the minimal binding region defined by deletion mutants and the effect of any point mutations, referenced to UniProtKB (2), the underlying protein sequence database. Protein interactions can be described down to the isoform level, or indeed to the post-translationally cleaved mature peptide level if such information is available in the publication, using the appropriate UniProtKB identifiers. The status of each of our proteins is checked with every release of UniProtKB—if a protein sequence has been withdrawn, the database is searched for a match (i.e. a transcript from the same gene, from the same organism and with >98% sequence identity) and the protein is remapped if possible. If a remapping is not possible, the sequence is retained within IntAct and can be accessed by users; a search for a match within UniProtKB is repeated with every new release. Similarly, with every release of UniProtKB, the sequence of every protein is checked and, if necessary, updated, with amino acid coordinates of interacting domains remapped to the updated sequence. While the vast majority of records within the IntAct molecular interaction database are annotated to the very detailed requirements of the curation rules agreed by the IMEx Consortium, a subset of records are annotated to the less-comprehensive MIMIx (3) standard. In practise, this means that while the details of the host organism, interaction and participant methodologies are recorded, as is the interaction directionality (e.g. bait/prey), the fine details of the construct are not. The data required by the user to ascertain confidence in a particular interaction evidence are, however, still captured in full. As from 2011, IMEx and MIMIx records are clearly differentiated within the database.
The IntAct database also captures protein–small molecule (including phospholipids), protein–nucleic acid and protein–gene loci interactions. In these cases the ChEBI (4), INSDC (5) and Ensembl/Ensembl Genomes (6,7) databases are the reference resources. A full set of curation rules has been developed for these interaction types, which are included within the IntAct curation rules published on the website (http://www.ebi.ac.uk/intact/site/doc/IntActAnnotationRules.pdf). IntAct has continued to contribute to the development of the PSI-MI controlled vocabularies (CV), which is referenced extensively throughout each database entry and added new terms relevant to these particular data types.
As of September 2011, IntAct contains 275 145 binary interaction evidences abstracted from 5009 scientific publications, referencing 57 857 proteins (as defined by UniProtKB), 144 small molecules (as defined by ChEBI) and 233 genes (as defined by Ensembl). It should be noted, that the phrase binary interactions does not necessarily relate to a direct interaction—the term also encompasses pairs of molecules which have artefactually been generated by the Spoke expansion model.
Quality control
Each entry in IntAct is peer reviewed by a senior curator, and not released until accepted by that curator. Additional rule-based checks are run at the database level, and manually fixed when necessary. Finally, on release of the data, the original author of each publication is contacted and asked to comment on the representation of their data; again manual updates are made to the entry should the author highlight any errors.
CONTRIBUTION TO IMEx
The IntAct molecular interaction database is a founder member of the IMEx Consortium, a collaboration of interaction databases that are working together to share annotation effort and produce a non-redundant set of experimental protein–protein interaction data, manually annotated to a consistent standard (S. Orchard et al., manuscript in preparation). To this end, all publications from a nominated set of journals are fully annotated to IMEx standards and both the publication, and the experimental evidences it contains, are allocated unique IMEx identifiers and made available on the IMEx website. Data is available using IntAct's PSICQUIC service (8), in addition to being made searchable on the IntAct website. Agreed updates to the IMEx curation rules are incorporated into the IntAct curation rule set. All data which is directly submitted to IntAct, as part of the publication process, is issued with an IMEx identifier and will be made available on both websites as soon as the corresponding article is published. A major effort will be made in 2012 to both ensure that a larger proportion of new data is immediately made part of the IMEx data set and to issue identifiers to records which are part of our existing catalogue, but not yet available via IMEx. Implementation of a new publication tracker database, IMEx Central (https://imexcentral.org/icentral), into the IntAct editorial tool should be achieved by the end of 2011.
RELATIONSHIP WITH UNIPROTKB AND THE UNIPROT GENE ONTOLOGY ANNOTATION PROJECT
IntAct has maintained a close working relationship with both the UniProt consortium and the Gene Ontology annotation (GOA) project (9), exporting selected binary pairs out to both the Annotation Comment (CC) INTERACTION line of UniProtKB and to the GOA project. Previously, the decision whether to export a particular binary pair was based purely on the interaction detection method(s) used, and all n-ary data, i.e. complexes involving three or more participants, was discounted. The method by which this export decision was made, was recently updated to a simple scoring system. All binary interactions evidences in the IntAct database, including those generated by Spoke expansion of co-complex data, are clustered to produce a non-redundant set of protein pairs (R. C. Jimenez et al., manuscript in preparation). Each binary pair is then scored, using a simple addition of the cumulated value of a weighted score for the interaction detection method and the interaction type for each interaction evidence associated with that binary pair, as described using the PSI-MI CV terms. The scores are given in Table 1, all children of each given parent receives that score. Only experimental data is scored, inferred interactions, for example, would be excluded. Any low confidence data or data manually tagged by a curator for exclusion from the process, would not be scored. Isoforms and post-processed protein chains are regarded as distinct proteins for scoring purposes.
Table 1.
Numerical parameters used to score binary interactions based on their class of interactions detection method and interaction type
| Weighting | |
|---|---|
| Interaction detection method | |
| Biochemical | 3 |
| Biophysical | 3 |
| Protein complementation assay (PCA) | 2 |
| Imaging techniques | 0.6 |
| Interaction type | |
| Association | 1 |
| Physical association | 2 |
| Direct interaction | 5 |
| Colocalization | 0.2 |
Child terms of these classes inherit the same weight.
EXAMPLE
Protein A–Protein B
The interaction has been shown by a single yeast two-hybrid experiment and also by a coimmunoprecipitation in which it was identified as part of an affinity complex isolated from a cellular environment.
1 × Y2H (PCA) + Physical interaction = 2 + 2 = 4
1 × coimmunopreciptiation + Association = 3 + 1 = 4
Total score = 8
Once the interactions have been scored, a threshold value has been agreed upon. When the calculated score of a binary interaction is greater than this threshold, it is exported to UniProtKB/GOA. Additional rules ensure that any protein pair scoring above this threshold must also include at least one evidence of a binary pair, excluding spoke expanded data, before export to UniProtKB/GOA.
These criteria ensure that:
Only experimental data is used for making the decision to export the protein pair to UniProtKB/GOA as a true binary interacting pair. An author may submit a secondary data set derived from the experimental data but this will not be included in the calculation.
The export decision is always based on at least two pieces of experimental data. A single evidence cannot score highly enough to trigger an export and
An export cannot be triggered if the protein pair only ever co-occurs in larger complexes, there must be at least one evidence that the proteins are probably in physical contact.
While these rules mean that currently only a small proportion of the binary pairs within IntAct are exported to UniProtKB and GOA, we believe that conservatively selecting protein pairs with a high degree of probability that these physically interact is the best service we can offer to these databases. It is our intention to add the additional non-redundant set of publications annotated within the IMEx consortium to this process in 2012, which should result in a marked increase in the number of lines exported. As from 2011, the UniProt Consortium is contributing to the records held within IntAct by curating interaction data directly into the database. We are happy to offer both export services and curation facilities to other databases wishing to establish a similar relationship with both IntAct and the IMEx Consortium.
NEW EDITORIAL TOOL
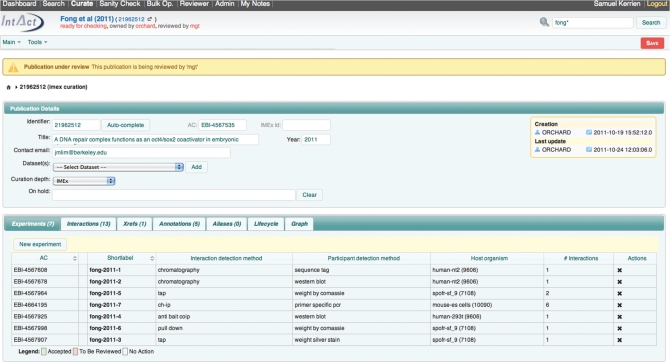
Manual interaction data curation is an arduous task that can be rendered more effective by using appropriate tools. With this in mind, we have redesigned our curation interface and streamlined the manual annotation process. The organization of this web-based curation tool reflects the complex nature of the underlying database structure, however provides easy navigation between connected entities such as publications, experiments, interactions and participating molecules. The interface facilitates MIMIx curation by providing all mandatory fields in a summary section for each entity while enabling curators to fill in more information in order to meet the more detailed requirement of IMEx curation (Figure 1). In order to facilitate communication during the entry quality control checking process, a publication lifecycle was designed and integrated into the heart of the application, thus significantly shortening the time to public release of curated records. New graphical components have been integrated in order to facilitate the interpretation of the data. A network visualization tool was added so that a single experiment as well as a complete publication can be viewed as a graphical network. Similarly, experimental features such as binding site and tags can be graphically displayed at the level of a given interaction, thus facilitating review by a senior curator. All data entities can be accessed via a REST URL, thus enhancing the accessibility of our curated data. Furthermore, we support the direct export of standard data formats such as PSI-MI XML 2.5 and PSIMITAB enabling curators to easily provide such file types to groups who submit data prior to publication. An administration console was also added to facilitate the work or senior curators and render the team less reliant on technical staff. This section comprises a user management system that enables a senior curator to easily create new user accounts and manage existing ones. The new IntAct curation interface is open source and can be freely used by third parties. We provide documentation on how to perform a local installation of the software (http://code.google.com/p/intact/).
Figure 1.
Screen shot of the new editorial tool.
The work of our curators will be further facilitated in the near future by integrating automated sanity checks on curated data and allow our team to identify curation issues faster. Furthermore, the curation interface will be closer integrated with IMEx Central to enhance communication with other IMEx partners and reduce the risk of redundant curation work. A number of external organization, InnateDB (10), I2D (11) and Molecular Connections (http://www.molecularconnections.com) are already collaborating with IntAct in order to use both the editorial tool and in-house quality control measures, to produce IMEx-level curated records, and other such collaborations are welcomed.
UPGRADED INTACT WEBSITE
The IntAct database is continuously growing and the scope of data captured is only getting broader. The IntAct public website has been updated to reflect these changes in data and improved visual components have been integrated. As the amount of data which can be displayed in any tabular display of interaction data increases, the ability to fit this onto a computer screen becomes more of a challenge. IntAct has responded to this by giving the user a choice of tabular visualization when the initial results of a search are displayed (Minimal, Basic, Standard, Expanded) with differing levels of detail immediately visible.
User-friendly inbound URLs
We have created simple URLs to access the molecular interactions in the IntAct database to allow clear linking from external resources. To access the details of a specific interaction, one can use the URL http://www.ebi.ac.uk/intact/interaction/<ACCESSION>. Alternatively, it is possible to access the results of a query using the URL http://www.ebi.ac.uk/intact/query/<QUERY>. It is planned for these URLs to be stable and not change with future updates of the website. Stable URLs to access other parts of the site are available on request.
Network visualization
In order to enhance the user experience when viewing molecular interaction, we have integrated CytoscapeWeb (12), an interactive network browser that we have customized to provide our users additional functionality such as edge merging to unclutter large networks, or different choices of graph layout. The ability to download data straight into the Cytoscape desktop application has been retained as CytoscapeWeb contains none of the plugin architecture functionality and users may wish to perform more complex analyses than is currently possible.
New export formats
In addition to the original PSI-MI XML and PSI-MITAB standard formats, we have added the possibility of exporting to BioPAX (13) levels 2 and 3 formats which is an RDF-based format widely used for the exchange of biological pathway data. RDF is a standard model for data interchange on the Web, being one of the technologies that empower the Semantic Web, a system that aims to help computers understand better the semantics of the provided data. To allow for more flexibility and freedom by the service consumers, other common RDF formats have been included as export options, such as RDF/XML and RDF/XML-ABBREV (RDF Syntax Recommendation), N3 (Tim Berners-Lee's Notation 3 Language), N-Triples (RDF Core's N-Triples Language) and Turtle (Terse RDF Triple Language). Following the Linked Data (14) principles, in the RDF output we have included dereferenceable URIs to identify the participant molecules in interactions and its cross-references, in order to improve the discovery of other related information on the Web. This further facilitates the inclusion of IntAct's data in the Semantic Web.
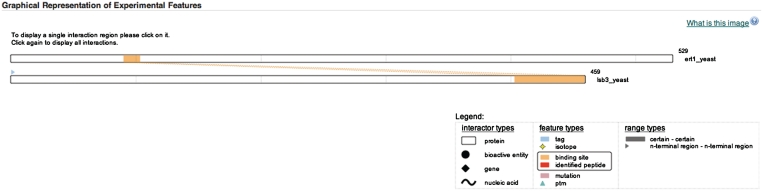
Experimental features such as binding site, tags, isotope labels, post-translational modifications, identified peptides and mutations are a valuable part of our IMEx-level manual curation which previously were only displayed textually in the interaction details. A new component has been designed and integrated to graphically represent this positional information on protein sequences (Figure 2). Participant proteins and features are displayed and scaled to represent the sequence length of the molecules. The user can interact with the graphical display to access additional experimental feature information and highlight interacting regions between proteins. Interactions with other molecule types, such as genes or small molecules are also visualized.
Figure 2.
New graphical component showing a representation of experimental and biological features available on the enhanced IntAct website.
FUTURE DEVELOPMENTS
Interaction confidence scoring within IntAct
The rules for export of interaction data to UniProtKB and GOA are not appropriate for visual representation, nor do they easily allow the external user to assess a ‘good’ interaction from one of low confidence as this score is simply additive and will continue to increase as further data on a specific binary pair enters the database. There are many systems available for scoring protein interaction data, based on various criteria including interaction evidences (15). To be able to systematically evaluate annotation evidences of individual interactions, we will soon implement MIscore, a confidence score based on common and minimum curated information reporting a molecular interaction experiment. MIscore relies on molecular interaction information compliant with the PSI-MI standards and annotated to at least MIMIx standards using the PSI-MI controlled vocabularies. For each binary pair of interacting partners, the experimental detection method, interaction type and the number of publications in which experimental evidences have been observed will be scored. The algorithm will then calculate a final normalized value between 0 and 1. In this way, the score will take into account the diversity of annotations reported for an interaction. As the method is linked to common curation standards and tools, this algorithm can be used not only to score, compare and assess interactions from IntAct but also interactions from other MIMIx-compliant databases and will work for any type of molecular interaction, not just PPIs. The normalization makes it easier for a user to understand the relevance of a particular score and enables easy filtering of low-scoring interactions out of a particular data set.
Enhanced graphical display of data
As described earlier, the use of CytoscapeWeb has provided IntAct with a visualization tool which can be maintained with low overhead but still allows a certain degree of customization to be built into the view. One such customization will be an interactive slider that will allow users to ‘fade out’ interactions by increasing the level of interaction confidence (as scored using MIscore) as they move the slider along a 0–1 scale. Downloads of the corresponding data will be available at any point in this process.
Quantitative data
The IntAct database is continually reviewing its ability to react to, and capture, new data types, as they are adopted by the community. Mass spectrometry-based affinity proteomics is now an increasingly popular technique for identifying molecular interactions. From such experiments, quantitative data, indicating not only which proteins are present in an interaction but also either their relative or absolute amounts within a complex, and changes in these amounts with changes in the cellular environment, can be generated. It will be a major challenge over the next 2–3 years to both capture and present such data in a way that database users can visualize the dynamic state of the macromolecular complex in question, and potentially also the functional consequences of such changes.
FUNDING
IntAct is funded by the European Commission under SLING, grant agreement number 226073 (Integrating Activity) within Research Infrastructures of the FP7, under PSIMEx, contract number FP7-HEALTH-2007-223411 and under APO-SYS, contract number FP7-HEALTH-2007-200767. Funding for open access charge: EMBL-EBI.
Conflict of interest statement. None declared.
REFERENCES
- 1.Kerrien S, Orchard S, Montecchi-Palazzi L, Aranda B, Quinn AF, Vinod N, Bader GD, Xenarios I, Wojcik J, Sherman D, et al. Broadening the horizon—level 2.5 of the HUPO-PSI format for molecular interactions. BMC Biol. 2007;5:44. doi: 10.1186/1741-7007-5-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.The UniProt Consortium. Ongoing and future developments at the Universal Protein Resource. Nucleic Acids Res. 2011;39:D214–D219. doi: 10.1093/nar/gkq1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Orchard S, Salwinski L, Kerrien S, Montecchi-Palazzi L, Oesterheld M, Stümpflen V, Ceol A, Chatr-aryamontri A, Armstrong J, Woollard P, et al. The minimum information required for reporting a molecular interaction experiment (MIMIx) Nat. Biotechnol. 2007;25:894–898. doi: 10.1038/nbt1324. [DOI] [PubMed] [Google Scholar]
- 4.de Matos P, Alcántara R, Dekker A, Ennis M, Hastings J, Haug K, Spiteri I, Turner S, Steinbeck C. Chemical entities of biological interest: an update. Nucleic Acids Res. 2010;38:D249–D254. doi: 10.1093/nar/gkp886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cochrane G, Karsch-Mizrachi I, Nakamura Y, International Nucleotide Sequence Database Collaboration The International Nucleotide Sequence Database Collaboration. Nucleic Acids Res. 2011;39:D15–D18. doi: 10.1093/nar/gkq1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Flicek P, Amode MR, Barrell D, Beal K, Brent S, Chen Y, Clapham P, Coates G, Fairley S, Fitzgerald S, et al. Ensembl 2011. Nucleic Acids Res. 2011;39:D800–D806. doi: 10.1093/nar/gkq1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kersey PJ, Lawson D, Birney E, Derwent PS, Haimel M, Herrero J, Keenan S, Kerhornou A, Koscielny G, Kähäri A, et al. Ensembl Genomes: extending Ensembl across the taxonomic space. Nucleic Acids Res. 2010;38:D563–D569. doi: 10.1093/nar/gkp871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aranda B, Blankenburg H, Kerrien S, Brinkman FS, Ceol A, Chautard E, Dana JM, De Las Rivas J, Dumousseau M, Galeota E, et al. PSICQUIC and PSISCORE: accessing and scoring molecular interactions. Nat. Methods. 2011;8:528–529. doi: 10.1038/nmeth.1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barrell D, Dimmer E, Huntley RP, Binns D, O'Donovan C, Apweiler R. The GOA database in 2009–an integrated Gene Ontology Annotation resource. Nucleic Acids Res. 2009;37:D396–D403. doi: 10.1093/nar/gkn803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lynn DJ, Winsor GL, Chan C, Richard N, Laird MR, Barsky A, Gardy JL, Roche FM, Chan TH, Shah N, et al. InnateDB: facilitating systems-level analyses of the mammalian innate immune response. Mol. Syst. Biol. 2008;4:218. doi: 10.1038/msb.2008.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Niu Y, Otasek D, Jurisica I. Evaluation of linguistic features useful in extraction of interactions from PubMed; application to annotating known, high-throughput and predicted interactions in I2D. Bioinformatics. 2010;26:111–119. doi: 10.1093/bioinformatics/btp602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lopes CT, Franz M, Kazi F, Donaldson SL, Morris Q, Bader GD. Cytoscape Web: an interactive web-based network browser. Nat. Immunol. 2010;26:2347–2348. doi: 10.1093/bioinformatics/btq430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Demir E, Cary MP, Paley S, Fukuda K, Lemer C, Vastrik I, Wu G, D'Eustachio P, Schaefer C, Luciano J, et al. The BioPAX community standard for pathway data sharing. Nat. Biotechnol. 2010;28:935–942. doi: 10.1038/nbt.1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bizer C. The emerging web of linked data. IEEE Intelligent Systems. 2009;24:87–92. doi: 10.1109/MIS.2009.102. [Google Scholar]
- 15.Ceol A, Chatr-Aryamontri A, Licata L, Peluso D, Briganti L, Perfetto L, Castagnoli L, Cesareni G. MINT, the molecular interaction database: 2009 update. Nucleic Acids Res. 2010;38:532–539. doi: 10.1093/nar/gkp983. [DOI] [PMC free article] [PubMed] [Google Scholar]