Abstract
SABIO-RK (http://sabio.h-its.org/) is a web-accessible database storing comprehensive information about biochemical reactions and their kinetic properties. SABIO-RK offers standardized data manually extracted from the literature and data directly submitted from lab experiments. The database content includes kinetic parameters in relation to biochemical reactions and their biological sources with no restriction on any particular set of organisms. Additionally, kinetic rate laws and corresponding equations as well as experimental conditions are represented. All the data are manually curated and annotated by biological experts, supported by automated consistency checks. SABIO-RK can be accessed via web-based user interfaces or automatically via web services that allow direct data access by other tools. Both interfaces support the export of the data together with its annotations in SBML (Systems Biology Markup Language), e.g. for import in modelling tools.
INTRODUCTION
The systematic study of complex interactions in biological systems requires detailed qualitative and quantitative information about single biochemical reactions in order to understand better the entirety of processes that happen in a biological system. For the quantitative analysis of biochemical reactions by modelling their enzyme kinetics, reliable kinetic data for the individual reaction steps are essential. Kinetic laws describing the dynamics of the reactions with their respective parameters determined under certain experimental conditions are mainly found in the literature. SABIO-RK (1,2) was developed as a database to store and structure kinetic data of biochemical reactions and their related information to support modellers and wet-lab scientists in understanding complex biochemical networks. The curation of all data in the database is used to achieve correctness and consistency within the database. Compared to other existing databases containing information about biochemical reaction kinetics [BRENDA (3), UniProt (4), BioModels (5), JWS Online (6)] that either are enzyme, protein or model databases SABIO-RK comprises a reaction-oriented representation of quantitative information on reaction dynamics based on a given selected publication. This comprises all available kinetic parameters together with their corresponding rate equations, as well as kinetic law and parameter types and experimental and environmental conditions under which the kinetic data were determined. Additionally, SABIO-RK contains information about the underlying biochemical reactions and pathways including their reaction participants, cellular location and detailed information about the enzyme proteins catalysing the reactions including the biological source.
At the beginning of the database development data were solely manually extracted from published literature. Now SABIO-RK constitutes an integration platform that supports the bundling of data inserted from literature, as well as directly submitted from lab experiments. In this process data which are in the pre-published phase are hidden for public access.
By the implementation of a new user interface and new web services the database now exhibits higher performance and more flexibility. Search criteria now include the search for organism taxonomy based on NCBI (7), compound classification based on ChEBI ontology (8) and tissue ontology based on BRENDA tissue ontology (BTO) (9) offering higher and more flexible usability of the database.
DATA CONTENT
For establishing a broad information basis SABIO-RK integrates data from different data sources. Mainly available information is extracted manually from literature which includes reactions, their participants (substrates, products), modifiers (inhibitors, activators, cofactors), catalyst details (e.g. EC enzyme classification, protein complex composition, wild-type/mutant information), kinetic parameters together with corresponding rate equation, biological sources (organism, tissue, cell location), environmental conditions (pH, temperature, buffer) and reference details. Data are adapted, normalized and annotated to controlled vocabularies, ontologies and external data sources. Additionally information about reactions and compounds are regularly updated with information from the KEGG database (10).
Detailed information about the protein catalysing a reaction is stored including information about specific isozymes or mutations used in the experiments, UniProt accession numbers and details about the composition of subunits forming the active enzyme. For some publications containing details about the reaction mechanism SABIO-RK also covers data about the elementary steps. This not only includes the kinetic data for the single elementary steps but also a graphical representation of the reaction mechanism.
Extracted information from literature is entered into a temporary database by using a web-based input interface. SABIO-RK always refers to the original source of kinetic data whereas values referring to a referenced paper are not linked to this publication. Before transferring the data to the final database, they are checked, complemented and verified by a curation team of biological experts to eliminate possible errors and inconsistencies.
Most of the publications are selected by reaction kinetics related keyword search in the PubMed database (7) or offered by collaboration partners. The selection of papers is not restricted to any organism or organism class. Yet there is a certain focus on data useful for collaborative Systems Biology projects in which we participate such as the Virtual Liver Network (http://www.virtual-liver.de/) or SysMO-LAB (Comparative Systems Biology of Lactic Acid Bacteria) (http://www.sysmo.net/).
As of October 2011, data from over 3400 publications have been curated and are stored in the database. Usually, one publication results in several entries if different reactions, enzymes, environmental conditions etc. are described. On average there are 10 entries per publication. An entry is a dataset which describes the outcome of a single experiment pertaining to one biochemical reaction. More specifically, it contains kinetic parameters measured under defined assay conditions and if available the corresponding kinetic law type and rate equation of the reaction catalysed by an enzyme derived from a specific organism. Currently SABIO-RK contains more than 42 000 curated single entries, for example ∼38% of them are related to the kinetic law type ‘Michaelis-Menten’, >14% of the entries contain diverse types of inhibitions, and ∼25% of the entries have no kinetic law type based on missing information in the publications. Kinetic parameters in SABIO-RK include more than 27 300 velocity constants (Vmax, kcat, rate constants), more than 30 900 Km values (including S_half for Hill equations) and about 7300 inhibition constants (Ki, IC50).
Kinetic data are available for about 660 organisms, 7300 different reactions and almost 1000 enzymes catalysing these reactions. About 2300 reactions containing kinetic data are linked to the reaction page of the KEGG LIGAND database. Two-thirds of the reactions are additional alternative reactions extracted from the publications for which kinetic data are available. Table 1 represents more detailed statistics for the ten most frequent organisms based on the number of entries in the database.
Table 1.
SABIO-RK statistics for the top 10 organisms
| Organism | Entries (total) | Mutant entries | Reactions (distinct) | EC numbers (distinct) | Velocity constants | Km values | Rate equations |
|---|---|---|---|---|---|---|---|
| Homo sapiens | 7028 | 2156 | 1233 | 348 | 5353 | 5757 | 3738 |
| Rattus norvegicus | 4286 | 759 | 821 | 285 | 2723 | 3188 | 2178 |
| Escherichia coli | 3828 | 1912 | 577 | 188 | 2976 | 3155 | 1988 |
| Saccharomyces cerevisiae | 1518 | 451 | 220 | 76 | 1044 | 1300 | 805 |
| Bos taurus | 982 | 59 | 276 | 86 | 579 | 722 | 471 |
| Oryctolagus cuniculus | 849 | 125 | 180 | 50 | 450 | 626 | 322 |
| Sus scrofa | 842 | 295 | 158 | 68 | 669 | 434 | 750 |
| Mus musculus | 809 | 92 | 236 | 92 | 456 | 290 | 603 |
| Enterococcus faecalis | 538 | 42 | 155 | 51 | 320 | 157 | 478 |
| Gallus gallus | 487 | 167 | 78 | 35 | 405 | 521 | 217 |
DATA INPUT AND CURATION
There are two different ways to insert kinetic data into SABIO-RK. Literature-based information is inserted by students and biological experts using a web-based input interface (11). More recently, data from lab experiments can be directly submitted and incorporated into SABIO-RK using a submission interface that accepts data described in the XML-based SabioML format. Data received in this format is directly inserted into the SABIO-RK database. This helps to automatize the submission process and provides a direct feed of kinetic data from the lab bench to the database, speeding up database population. Together with collaboration partners we have developed a tool for capturing, analysis and submission of data based on this interface (12), which is used for data submission of high-throughput kinetic assay results performed by collaboration partners in Manchester.
The web-based input interface was developed to enable the input of literature data into SABIO-RK. This interface is password protected and is used by our students and collaboration partners to insert their literature data first into a temporary database. The interface consists of several web-pages with form fields and selection lists for structured data input. The same interface is also used for the curation process. It implements a variety of constraints starting from simple data format checks like validation of numbers, to sophisticated tasks to avoid errors and inconsistencies. This includes e.g. checking if all parameters in a kinetic law formula are in the list of kinetic parameters, if all compound-dependent parameter types are related to a chemical compound, and if all reaction participants are provided and have a defined role in the reaction equation as well. In order to represent consistent biochemical reactions their equations are automatically generated from the list of substrates and products and cannot be changed manually.
For consistency and maintenance of the controlled vocabulary, and to avoid duplicate entries, lists of compounds, reactions, organisms, tissues, cellular locations, kinetic law types, parameter types and units already existing in the SABIO-RK database are provided as selection lists or can be searched in the input interface. The controlled vocabularies used for these lists are generated by extracting terms from the following external sources: organism names from NCBI taxonomy (7), tissues and cellular locations from BRENDA (3), types of kinetic laws and parameters from Systems Biology Ontology (SBO) (13) and units from the International System of Units (SI, http://www.bipm.fr/en/si/). The term lists also contain synonyms referring to the same content to enable the search for alternative names of compounds etc. These controlled vocabularies together with annotations to external data resources and ontologies are used to identify and relate the data to their biological context. Biological ontologies used for annotations in SABIO-RK are ChEBI, SBO, BTO and NCBI taxonomy. A shared vocabulary and defined standards for storage, representation and export of data are important to avoid misinterpretations and to relate information to and exchange data with external sources. To unambiguously identify entities or terms and to facilitate search, interpretation and comparison of the data, SABIO-RK standardizes the data in a uniform structure.
Especially for unpublished data from lab experiments within collaboration projects SABIO-RK is able to define restricted access to the data. Within the database different rights are managed based on various group definitions. So users can enter their data into the database without making them visible to the public.
DATA ACCESS
To access the data in SABIO-RK web-based user interfaces and web-services are available offering the possibility of submitting complex searches by defining various search criteria. All interfaces support the export of the data together with its annotations in SBML format [Systems Biology Markup Language (14)]. SBML is a widely-used data exchange format in Systems Biology and thus well suited for exchanging the data with other tools, e.g. for its subsequent import in modelling tools for the setup of quantitative biochemical models for simulation.
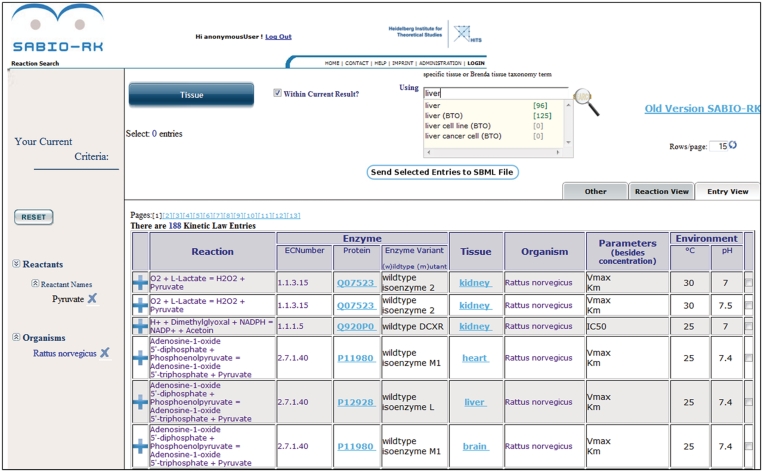
The web-based user interfaces enable the user to search for reactions and their kinetics by specifying characteristics of the reactions. It offers the creation of complex queries by specifying reactions by their participants (substrates, products, inhibitors, activators etc.) or identifiers (KEGG or SABIO-RK reaction identifiers and KEGG, SABIO-RK, ChEBI or PubChem (7) compound identifiers), pathways, enzymes, UniProt identifiers, organisms, tissues or cellular locations, kinetic parameters, environmental conditions or literature sources (Figure 1). Several search criteria (organism, tissue and reactant) are based on biological ontologies which define controlled vocabularies and relations between objects. These ontologies offer various levels of classification, which can be used in the search, relaxing or restricting it. Hierarchies built based on the ontological relations are implemented in the SABIO-RK search options for advanced functionality of the database. The search for organisms can be extended by the search for organism classifications based on the NCBI taxonomy, e.g. search for ‘Mammalia (NCBI)’. The tissue search in SABIO-RK includes the possibility to use BRENDA Tissue Ontology terms, e.g. search for ‘liver (BTO)’ offering more liver related entries compared to simple ‘liver’ search (Figure 2). And for a more comprehensive search for chemical compounds the is_a relationships extracted from the ChEBI Ontology are included in the database search options for reaction participants.
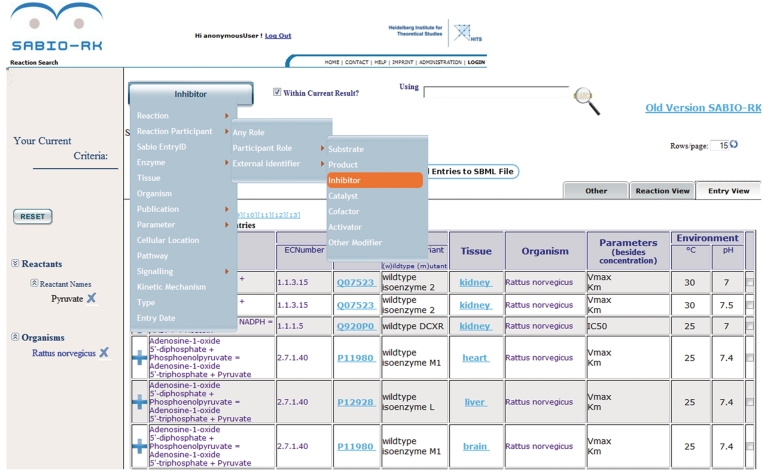
Figure 1.
SABIO-RK web interface screenshot using the drop-down menu to select search criteria (e.g. inhibitor).
Figure 2.
SABIO-RK web interface screenshot of query results in entry view (e.g. search for reactions containing Pyruvate in Rattus norvegicus).
Based on comprehensive annotations of data in SABIO-RK links to other databases and ontologies are included enabling the user to obtain further details, for example about reactions, compounds, enzymes, proteins, tissues, or organisms. On the other hand external databases offer their users the access to reaction-based kinetic data in SABIO-RK via cross-references. ChEBI compounds participating in reactions as substrates or products are linked to SABIO-RK reactions in the cross-references field ‘Reactions & Pathways’. KEGG implemented the links to SABIO-RK reactions from KEGG LIGAND reaction pages. Kinetic data entry details and corresponding annotations to external databases and ontologies can be exported with the data from SABIO-RK in SBML, compliant with the MIRIAM standard (15). For later tracking of the original data source SABIO-RK reaction and kinetic law identifier are themselves listed as MIRIAM data types.
Currently SABIO-RK has two different versions of web-based user interface, due to a transition process. The ‘old’ user interface had as a main shortcoming that only after sending a query the user gets an answer if data meeting the search criteria are present in the database. We wanted to be able to show the user beforehand how many results can be expected for a given set of search criteria. To this end, we had to improve the search efficiency. We changed from an SQL-based search mechanism to inverted indexing for all data entries (16). This inverted indexing technique is used in a new web interface developed for SABIO-RK. Now users can see the number of kinetic data entries for a given query while formulating the query and before searching the database. This improves the usability of the web interface and supports the users’ search behaviour. For the representation of the search results the user can select two different overview result lists: (i) the Entry View (Figure 2) shows the list of entries with general information including reaction equation, enzyme details, tissue, organism, parameters and environmental conditions; (ii) the Reaction View (Figure 3) groups the entries based on SABIO-RK reactions and shows corresponding KEGG reaction identifiers. Additionally reaction related information visualization is offered representing the relation between a reaction and corresponding enzymes, organisms and tissues. More detailed information within single database entries can be displayed by expanding the overview information available in both views (Figure 4). Enabling the checkbox in the overview offers the selection of entries for data export.
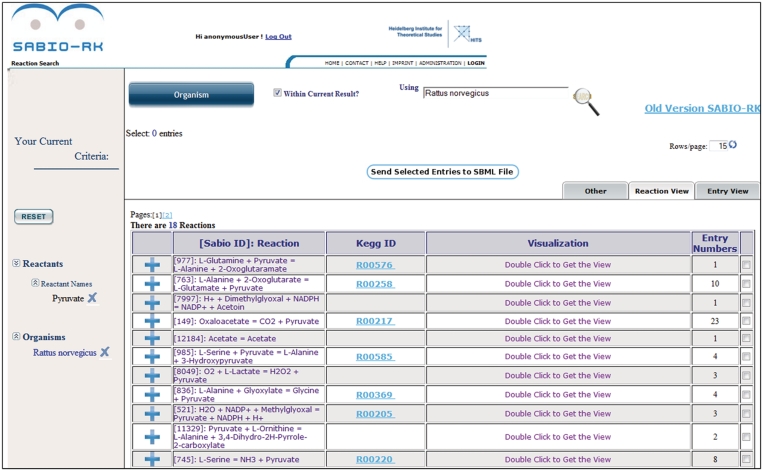
Figure 3.
SABIO-RK web interface screenshot of query results in reaction view (e.g. search for reactions containing Pyruvate in Rattus norvegicus).
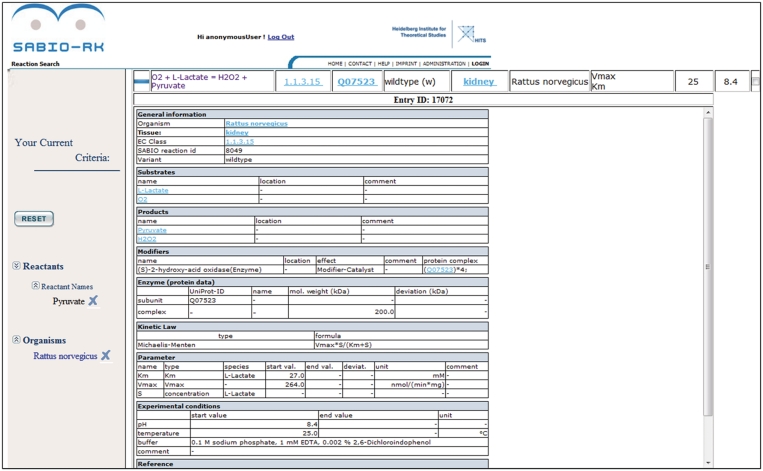
Figure 4.
SABIO-RK web interface screenshot of a single database entry (e.g. entry ID 17072).
For programmatic access to the database additionally to the already existing SOAP-based web services new web services were implemented allowing data access via HTTP requests following a Representational State Transfer (REST) approach (17). This new approach is much more similar to the web user interface, thus making it easy to first test queries in the web user interface and then turning them into a web service consumer. For the definition of queries simple URLs could be defined including request parameters. Entries can be requested directly by using the database entry ID or can be searched using the same search options available in the web-based user interfaces. The output of this web services search is in XML or SBML format. With the different types of programmatic access SABIO-RK facilitates the exchange of kinetic data between experimentalists and modellers and supports the integration of automatic data access in applications e.g. the systems biology modelling platforms CellDesigner (18) and SYCAMORE (19).
SUMMARY
SABIO-RK is a curated database containing biochemical reactions and theirs kinetics. It supports experimentalists and modellers of biochemical networks to obtain and compare data about reactions, their kinetics and related information like for example cellular locations, tissues, and organisms. The kinetic information is either manually extracted from literature or directly submitted from lab experiments. The annotation to controlled vocabularies, ontologies and external databases allows complex searches in the database, linking to external sources and the comparison of data. A new developed web-based user interface and new RESTful web services offer a much faster and more convenient access to the database compared to the old applications. Some improvements in the search functionality like easy combination of attributes are in progress. Complex datasets can be exported from SABIO-RK in SBML format for further processing and integration. Currently we are working on expanding the data export functionality including table and BioPAX formats.
FUNDING
Klaus Tschira Foundation (http://www.klaus-tschira-stiftung.de/); the German Federal Ministry of Education and Research (http://www.bmbf.de/) through Virtual Liver and SysMO-LAB (Systems Biology of Microorganisms), and the DFG LIS (http://www.dfg.de/), under the short title ‘Integrated Immunoblot Environment’. Funding for open acess charge: HITS gGmbH (http://www.h-its.org).
REFERENCES
- 1.Wittig U, Golebiewski M, Kania R, Krebs O, Mir S, Weidemann A, Anstein S, Saric J, Rojas I. Proceedings of the 3rd International workshop on data integration in the life sciences (DILS'06), Hinxton, UK. Lecture Notes in Computer Science, 4075. Berlin und Heidelberg: Springer; 2006. SABIO-RK: integration and curation of reaction kinetics data; pp. 94–103. [Google Scholar]
- 2.Rojas I, Golebiewski M, Kania R, Krebs O, Mir S, Weidemann A, Wittig U. Storing and annotating of kinetic data. In Silico Biol. 2007;7(Suppl 2):37–44. [PubMed] [Google Scholar]
- 3.Scheer M, Grote A, Chang A, Schomburg I, Munaretto C, Rother M, Söhngen C, Stelzer M, Thiele J, Schomburg D. BRENDA, the enzyme information system in 2011. Nucleic Acids Res. 2011;39:D670–D676. doi: 10.1093/nar/gkq1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.The UniProt Consortium. Ongoing and future developments at the Universal Protein Resource. Nucleic Acids Res. 2011;39:D214–D219. doi: 10.1093/nar/gkq1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Le Novère N, Bornstein B, Broicher A, Courtot M, Donizelli M, Dharuri H, Li L, Sauro H, Schilstra M, Shapiro B, et al. BioModels database: a free, centralized database of curated, published, quantitative kinetic models of biochemical and cellular systems. Nucleic Acids Res. 2006;34:D689–D691. doi: 10.1093/nar/gkj092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Olivier BG, Snoep JL. Web-based kinetic modelling using JWS Online. Bioinformatics. 2004;20:2143–2144. doi: 10.1093/bioinformatics/bth200. [DOI] [PubMed] [Google Scholar]
- 7.Sayers EW, Barrett T, Benson DA, Bolton E, Bryant SH, Canese K, Chetvernin V, Church DM, DiCuccio M, Federhen S, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2011;39:D38–D51. doi: 10.1093/nar/gkq1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Matos P, Alcántara R, Dekker A, Ennis M, Hastings J, Haug K, Spiteri I, Turner S, Steinbeck C. Chemical entities of biological interest: an update. Nucleic Acids Res. 2010;38:D249–D254. doi: 10.1093/nar/gkp886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gremse M, Chang A, Schomburg I, Grote A, Scheer M, Ebeling C, Schomburg D. The BRENDA Tissue Ontology (BTO): the first all-integrating ontology of all organisms for enzyme sources. Nucleic Acids Res. 2011;39:D507–D513. doi: 10.1093/nar/gkq968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kanehisa M, Goto S, Furumichi M, Tanabe M, Hirakawa M. KEGG for representation and analysis of molecular networks involving diseases and drugs. Nucleic Acids Res. 2010;38:D355–D360. doi: 10.1093/nar/gkp896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rojas-Muijca I, Golebiewski M, Kania R, Krebs O, Mir S, Weidemann A, Wittig U. Proceedings of the 2nd International Symposium on ‘Experimental Standard Conditions of Enzyme Characterizations’, Ruedesheim am Rhein, Germany. Berlin: Logos; 2007. SABIO-RK (System for the Analysis of Biochemical Pathways Reaction Kinetics) pp. 189–202. [Google Scholar]
- 12.Swainston N, Golebiewski M, Messiha ML, Malys N, Kania R, Kengne S, Krebs O, Mir S, Sauer-Danzwith H, Smallbone K, et al. Enzyme kinetics informatics: from instrument to browser. FEBS J. 2010;277:3769–3779. doi: 10.1111/j.1742-4658.2010.07778.x. [DOI] [PubMed] [Google Scholar]
- 13.Le Novère N. Model storage, exchange and integration. BMC Neurosci. 2006;7(Suppl 1):S11. doi: 10.1186/1471-2202-7-S1-S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hucka M, Finney A, Sauro HM, Bolouri H, Doyle JC, Kitano H, Arkin AP, Bornstein BJ, Bray D, Cornish-Bowden A, et al. The systems biology markup language (SBML): a medium for representation and exchange of biochemical network models. Bioinformatics. 2003;19:524–531. doi: 10.1093/bioinformatics/btg015. [DOI] [PubMed] [Google Scholar]
- 15.Le Novère N, Finney A, Hucka M, Bhalla US, Campagne F, Collado-Vides J, Crampin EJ, Halstead M, Klipp E, Mendes P, et al. Minimum information required in the annotation of models (MIRIAM) Nat. Biotechnol. 2005;23:1509–1515. doi: 10.1038/nbt1156. [DOI] [PubMed] [Google Scholar]
- 16.Shi L, Jong L, Mueller W, Algaa E. 50x Faster: Speeding up an SQL-based legacy system with few changes. Lecture Notes in Informatics (LNI) - Proceedings, Series of the Gesellschaft für Informatik (GI) 2011;Vol. P-192 [Google Scholar]
- 17.Richardson L, Ruby S. Restful web services. 1st edn. Sebastopol, CA: O-Reilly Media; 2007. [Google Scholar]
- 18.Funahashi A, Jouraku A, Matsuoka Y, Kitano H. Integration of CellDesigner and SABIO-RK. In Silico Biol. 2007;7:S81–S90. [PubMed] [Google Scholar]
- 19.Weidemann A, Richter S, Stein M, Sahle S, Gauges R, Gabdoulline R, Surovtsova I, Semmelrock N, Besson B, Rojas I, et al. SYCAMORE - A SYstems biology Computational and Analysis MOdeling Research Environment. Bioinformatics. 2008;24:63–64. doi: 10.1093/bioinformatics/btn207. [DOI] [PubMed] [Google Scholar]