Abstract
Minimotif Miner (MnM available at http://minimotifminer.org or http://mnm.engr.uconn.edu) is an online database for identifying new minimotifs in protein queries. Minimotifs are short contiguous peptide sequences that have a known function in at least one protein. Here we report the third release of the MnM database which has now grown 60-fold to approximately 300 000 minimotifs. Since short minimotifs are by their nature not very complex we also summarize a new set of false-positive filters and linear regression scoring that vastly enhance minimotif prediction accuracy on a test data set. This online database can be used to predict new functions in proteins and causes of disease.
INTRODUCTION
A common theme in protein activity regulation is the binding of a structural domain of one protein to a short, contiguous peptide segment of another. From a bioinformatics perspective, identifying domain signatures has been incredibly useful in formulating hypotheses regarding the biological function of otherwise uncharacterized proteins. The success of such methods is due in part to the high sequence complexity of these relatively large domains (approximately 100 residues in length), as well as their common evolutionary heritage, which allow for high-confidence domain identification with few false positives. The short, contiguous segments [termed minimotifs or short linear motifs (SLiMs)] are just as useful in identifying the roles of proteins, but are more difficult to identify with high accuracy. Nevertheless, several bioinformatics resources exist for querying protein sequences for the existence of minimotifs, including Minimotif Miner (MnM), the Eukaryotic Linear Motif (ELM) resource and other specialized databases (1–10). It remains an ongoing pursuit to increase both the sensitivity and accuracy of minimotif prediction in proteins.
This article summarizes the latest release and developments of the MnM database and webserver, version 3.0; additional details can be found in the new MnM user guide on the MnM website. Efforts since our last release in 2008 (4) have concentrated on two fronts: improved filters which increase the accuracy of minimotif prediction by removing false positives (11–13), and increasing the size of the MnM database through both manual annotation of minimotifs from the literature and federation with other databases including PhosphoSite, DOMINO, MEROPS, UniProt, PepX, 3DID, PeptiDB and HPRD (14–21). MnM 3 now includes a total of 294 933 minimotif definitions, consisting of 880 consensus minimotifs and 294 053 instances. These minimotifs span three biological activities: trafficking, binding and modifying. Multiple filters have been introduced since our 2008 release of MnM 2, the most important being a combined filtering approach that can result in 90% accuracy of minimotif prediction with few false positives using one scoring threshold or even 38% identification of minimotifs with no false positives with a more stringent threshold (submitted for publication). The score used by this combined filter is now used as the default ranking of the minimotif list, rather than the frequency score used in MnM 2.
Besides their role in recognition by protein domains, minimotifs have a number of important biological roles. In addition to binding, minimotifs are often determinants for post-translational modifications and trafficking proteins to specific parts of cells. Minimotifs are also involved in cell signaling and regulation (22,23). A number of minimotifs are mutated in different disease and pathogens such as viruses tend to exploit host machinery by viral encoded minimotifs (24–26). Due to their role in disease, the actions of several drugs are based on a minimotif-mimetic mechanism (27,28).
RESULTS
Revised minimotif model and new entries in Minimotif Miner 3.0
Prior to adding minimotifs to the MnM 2 database, we first reevaluated our previous model, which presented 22 attributes of a minimotif (12). We have now revised this model to include 28 attributes as shown in Figure 1. This model contains a protein sequence definition and a functional definition where the sequence definition describes the chemistry of the motif. The sequence definition can be an instance or a consensus sequence. Instances are the exact amino acid sequence found in the protein that contains the minimotif; whereas, a consensus sequence is an interpretation of a set of instances that indicates degeneracies at certain positions in the amino acid sequence. The consensus sequence definition format is largely based on that previously proposed by the Seefeld Convention and later modified for MnM (12,29). These modifications include an extensible expanded definition of the covalent chemistry of the minimotif containing the position within the protein, any modified residues and their position in the sequence, and a description of any post-translational modifications of amino acids in the sequence and corresponding accession numbers from the Psi-Mod database (30).
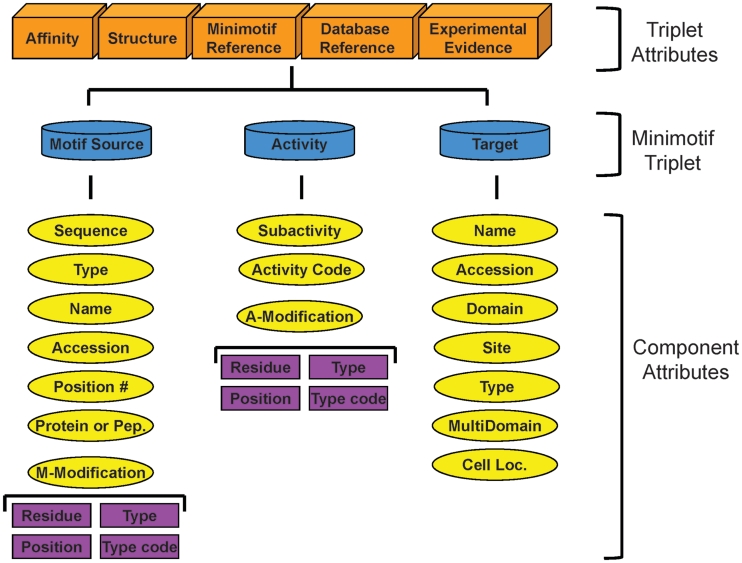
Figure 1.
Revised minimotif model. The key elements of the minimotif syntax are colored blue. Orange boxes indicate attributes that are unique to specific minimotif triplets. Yellow ovals are for different attributes of minimotif triplet elements. All attributes except those in the purple boxes were previously described in our minimotif model and the purple boxes are new attributes to define motif modifications and activity modifications (12).
The functional component of the minimotif model is centered around a syntactical triplet where the motif source is the subject, the activity is the verb and the target that engages the minimotif is the object. There are unique properties to this triplet such as an affinity, structure, minimotif reference, database reference for cross referencing external databases and experimental evidence that support the minimotif.
The motif source, activity and target have a number of attributes previously modeled, but here we have renamed the ‘required modification’ to ‘motif modification’ to better distinguish this attribute from ‘activity modification’. Motif modification is when a motif needs to be covalently modified to engage the target such as when a minimotif must be phosphorylated to bind 14–3–3. Whereas, an activity modification describes a situation where the target is an enzyme that covalently modifies the minimotif as when a minimotif becomes myristoylated. The description of these modifications requires more detail than in our original model. To accurately describe these modifications the new model includes the ‘residue’ that is modified, the sequence ‘position number’ of the sequence, the ‘type’ of modification and the ‘type code’ number of the modification, which for the most part makes use of accession ids in the Psi-Mod database (30).
Since the release of MnM 2, the total number of minimotif sequences has increased from about 5300 to almost 300 000 (Table 1) (4). The majority of these new entries have been gleaned from federation with other open databases and some were manually annotated from the primary literature using the MimoSA annotation helper application designed for this task (31). All minimotif entries are annotated using our revised minimotif syntax model (Figure 1) (12). The majority of growth comes from addition of instances. We have focused on instances because the Minimotif Miner query engine can be used to generate consensus sequence from any set of instances (12). Most of the minimotifs added from external sources are for post-translation modifications. Minimotifs are found in approximately 50 000 different proteins in many different species; most minimotifs are for mammalian organisms, although MnM does contain some bacterial, yeast and invertebrate minimotifs as well. The number of domains that interact with or associate with minimotifs in MnM is approximately 2600 suggesting that there are still many minimotifs yet to be discovered.
Table 1.
Growth of minimotif entries in MnM
| Category | MnM | MnM 2 | MnM 3 |
|---|---|---|---|
| Total | |||
| Motif sequences | 462 | 5089 | 294 933 |
| Consensus sequences | 312 | 858 | 880 |
| Instance sequences | 44 | 4229 | 294 053 |
| Post-translational modifications | 116 | 663 | 210 949 |
| Binding | 162 | 4689 | 4922 |
| Trafficking | 34 | 195 | 228 |
| Required for cell process | – | – | 47 |
| Unique | |||
| Motif sequences | 312 | 2224 | 185 833 |
| Motif proteins | <312 | 1211 | 49 671 |
| Motif targets | <312 | 687 | 2620 |
There is a minimal set of attributes necessary to define a minimotif for entry into MnM. The minimotif sequence types are classified as either an instance or a consensus sequence, which each have a minimal set of attributes. Consensus sequence definitions must have an amino acid sequence less than 15 residues long, activity and subactivity, literature reference, one or more experimental techniques in support of the minimotif, and annotation of any post-translational modifications to the minimotif sequence (residue modified, position in sequence, type of modification and Psi-Mod id for the modification, if available). Instances contain this attribute set, but also must have a name of the sequence harboring the minimotif, whether the source protein is a peptide fragment or a protein, and if a protein, must have an accession number to one of the available protein databases. While we prefer to have information about the target molecule that is associated with the minimotif, this is not required in the minimal set because there is value for such database entries in that this information can be used to identify unknown targets by mining-based approaches. For example, an instance of a phosphorylation site on a protein substrate can be used with kinase consensus sequences in the database to predict the target kinase. The 28 attributes of minimotifs are stored in a MySQL database. For the approximately 6000 manually annotated minimotifs, all 28 attributes were entered, except in the cases where information was not available from the literature. For example, some minimotifs do not have structures or affinities. We note that many of the minimotifs imported from external databases have the minimal set of information required to define a minimotif, but are often missing many of the other attributes defined in our model; we only imported minimotifs that have the minimal set of attributes.
Minimotif filtering to reduce false-positive predictions
The major difficulty in identifying functional minimotifs within a protein sequence of interest is the high false-positive rate—that is, a large number of predicted minimotifs do not perform the predicted biological function, but coincidentally share the minimotif sequence signature present in other biologically active proteins. These false-positive predictions are notoriously difficult to filter out based on sequence definitions alone, due to the inherently low-sequence complexity of minimotifs (7,32). However, additional context information (beyond amino acid sequence) can be used to narrow the search and effectively filter these false positives, increasing the accuracy of minimotif prediction (11,13). Such context information is routinely employed by individual researchers when evaluating minimotif prediction results. For instance, a researcher studying nuclear import in mouse neurons would quickly discard motif predictions regulating bacterial cell division. In this case, the researcher would be imparting context-specific information about molecular function, cellular function and taxonomy to rule out an obvious false positive. While effective, such a filtering technique is highly inefficient both in the time it takes an individual to prune the results list, as well as in the breadth of understanding required to effectively filter all false positives. Over the past 2 years, several contextual filters have been added to the MnM web service, which have been demonstrated to be highly effective in improving the accuracy of minimotif prediction thereby increasing the ease of interpretation of the Minimotif Miner results (11,13).
The original implementation of MnM 1.0 did not attempt to filter any false positives, but ranked minimotif predictions in descending order of sequence complexity. A scoring metric for location of a minimotif on a protein surface, and evolutionary conservation among divergent species was also provided. MnM 2 allowed the user to filter the results list based on particular minimotif activities of interest and also separated minimotif instances from consensus definitions. Neither of these functionalities formally removes false positives, they simply assist the user in filtering based on his/her own knowledge.
Our first step toward knowledge-based filtering was to model minimotif definitions in a richer way—formally modeling the source and target proteins (12). Tying the source/target protein information to the minimotif definition provides a relationship to the taxonomy of the observed activity, a relationship to other related species through homology databases/searches, a relationship to molecular and cellular function through the source/target annotation and the use of the Gene Ontology definitions, and a relationship to other proteins in the same biological pathway through protein–protein interaction databases (21,33–37). In this manner, the use of context-specific definitions can be applied to filter false-positives computationally, removing that burden from the user.
Molecular/cellular function
Knowing that the source protein and target interact directly is highly helpful in identifying true positives in a minimotif search, as minimotif activities require an interaction between the source and target. In the absence of such direct interaction, filtering for source/target pairs, which are active in the same molecular/cellular pathway can also be useful. Functions of source/target pairs are accessed from the Gene Ontology database, which allows for filtering based on the molecular/cellular function of the source/target pairs (38). This filtering technique can be used to restrict results to only source/target pairs, which share a common function, or can be extended to source/target pairs that share a related function. Three thresholds are provided in MnM 2.1 for varying the relatedness of the functions to be filtered (13). The best performing cellular function filter is estimated to result in ∼26% sensitivity with 6% selectivity for a combined discrimination ratio of 4.6 whereas the best performing molecular function filter has a discrimination ratio of 2.9 (Table 2). Sensitivity is the percentage of true positives that are not filtered out, whereas selectivity is the percentage of true negatives that are not removed by the filter (11,13). The discrimination ratio is sensitivity/selectivity.
Table 2.
Comparison of different minimotif filters
| Minimotif filter | Area under ROC curve | P-value | Discrimination ratio | Reference |
|---|---|---|---|---|
| Frequency score | 0.7 | 0.08 | ND | (11,13) |
| Cellular function | 0.7 | 0.12 | 4.6 | (13) |
| Cellular function + frequency score | 0.9 | 0.0002 | – | (13) |
| Molecular function | 0.8 | 0.03 | 2.9 | (13) |
| Molecular function + frequency score | 0.9 | 0.002 | – | (13) |
| Protein–protein interaction | 0.9 | 0.001 | 12.5 | (11) |
| Genetic interaction | – | – | 7.3 | Submitted for publication |
| Surface prediction filter | 0.3 | 1 | – | (7) |
| Multifilter | 0.94 | 9.7 e−278 | – | Submitted for publication |
Protein–protein interactions
MnM 2.2 allowed the user to filter results based on known protein–protein interaction (PPI) networks (11). The logic behind this filter is that minimotif predictions are filtered on the basis of experimental verification of the interaction between source and target proteins. MnM makes use of six external databases containing more than 300 000 non-redundant PPIs: DiP, Entrez Gene, HPRD, MINT, VirusMINT and IntAct (21,33–35,37,39). In the most stringent use of the PPI filter, only exact matches between source and target are reported, and the predicted minimotif represents a hypothetical mode of interaction for the known PPI. While effective, this stringent filter is limited due to the relatively small number of established PPIs. For this reason, the user can extend the filter to include homologous proteins for both the known source and known target of the PPI. This can be done in one of two ways: one, by accessing homologous protein clusters via the HomoloGene database; two, by using BLAST similarity searches to predict homologous proteins not included in HomoloGene (39). Ten default BLAST thresholds are provided in the motif filtering dialog box (Figure 2) accounting for a total of 12 possible PPI filtering choices. The base-level PPI filter is estimated to result in ∼62% sensitivity with 2% selectivity for a combined discrimination ratio of 29; this is the best performing filter and significantly reduces false positives (Table 2).
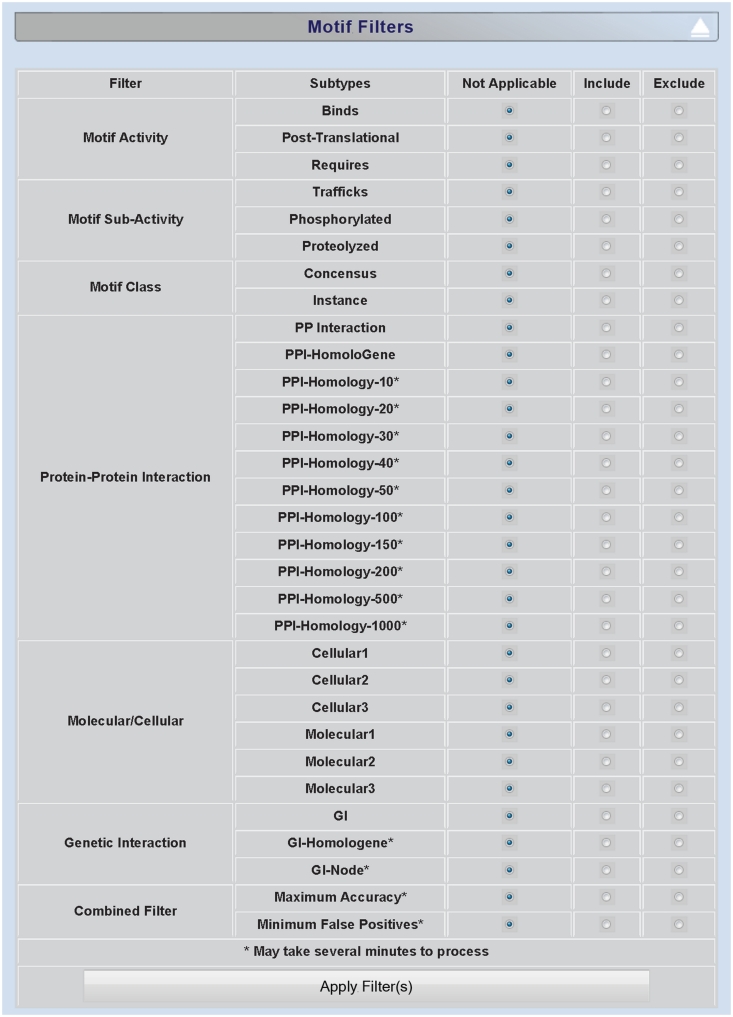
Figure 2.
Screenshot of minimotif filter selection page. Screenshot of MnM 3 filter section for choosing approaches for filtering out false-positive minimotifs.
Genetic interactions
A genetic interaction (GI) helps to identify that there is a functional relationship between two proteins. In some cases, this can be due to direct interactions or modifications of one protein by another. If a minimotif source and predicted target protein have a GI, this prediction can provide a mechanistic explanation for the observed relationship. Since the two proteins of a GI have this relationship, these proteins are more likely to have a minimotif than two unrelated proteins. This concept was implemented in three different GI filters on MnM 2.3 (submitted for publication). The basic GI filter identifies those motif/target pairs where there is a known GI and was the GI filter with the highest accuracy; the GI-node based filter extends the GIs for the sources and target an additional interaction away to a path length of 2; the GI-HomoloGene filter takes advantage of orthologous GIs. The basic GI filter had a discrimination ratio of 7.3, which was better than the GI-node and GI-HomoloGene filters with ratios of 4.5 and 2, respectively. The primary difference comes from a poorer selectivity in removing true negatives (∼3% versus ∼12%); similar sensitivities of 21% and 24% were observed for these filters. The basic GI filter also had a better discrimination ratio than the cellular and molecular function filters, but was not as high as the PPI filter (Table 2).
Combined filter approach
When examining the effectiveness of the cellular / molecular function filters, we discovered that combining two filters provided greater accuracy in minimotif prediction than either filter alone (13). We have recently extended this idea by training a linear combination of all the filters to maximize both the accuracy and specificity in minimotif search. Using this approach with one threshold, the resulting combined filter allows us to increase accuracy to 90%, while a more stringent threshold retained 39% of the true positives while rejecting all false positives in a large test data set. The elimination of all false positives represents an important milestone in minimotif prediction. MnM 3 provides access to all minimotif filters and now ranks all minimotif predictions using this new combined filter score. The results can also be filtered according to either the threshold for maximizing accuracy or for maximum stringency in removing all false positives (Figure 2). In the minimotif results table, minimotifs with scores above the threshold of 0.91 are highlighted green (produce no false positives on a test data set), between 0.24 and 0.90 are highlighted yellow (produce high recovery of true minimotifs with only 2% false positives on a test data set). Experimentally validated minimotifs are distinguished from predictions by highlighting the minimotifs blue in the results table.
Uses of Minimotif Miner
The major workflow in minimotif miner is to search a single protein query for the presence of minimotifs. This is geared toward identifying new functions in proteins, minimotif determinants of protein–protein interactions, or for matching post-translational modifications with potential enzymes that catalyze such modifications. Many different subsets of minimotifs can be selected by using the filtering section of the MnM results page (Figure 2). Once a custom filter combination is selected with radio buttons, selection of the ‘Apply Filter(s)’ button will repopulate the motif results table with the search results. As the filtering approaches have grown, this has now been moved from the bottom of the protein sequence menu to a separate expandable section on the results page.
To provide an example of the uses of different filters we explore a sample analysis of HIV-1 Nef, (NP_057857). We chose Nef because it is a well-studied protein and we could evaluate minimotif predictions using HIVToolbox (40). Although we expected better filtering from the new algorithm, only a small portion of known minimotifs have been identified and added to MnM, thus we would only expect that a subset of predicted minimotifs would have been previously experimentally validated. In the old MnM 2 output, the minimotifs were rank ordered by frequency score. This ordering often strongly selects for a high ranking of instances, which generally are far more information-rich than consensus sequences. The new ranking in MnM 3 depends on many types of different data and filter testing indicates that the new filters are superior to the minimotif ranking used in MnM 2.
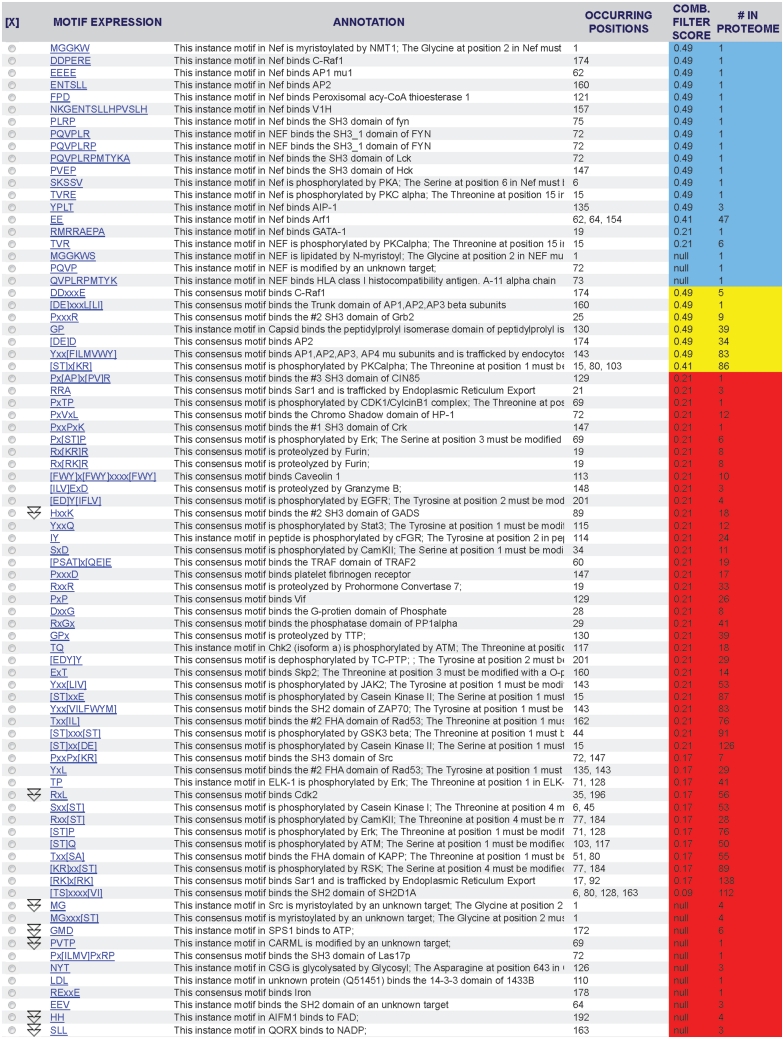
The new default filter ranks 21 minimotif predictions for Nef with a score between 0.24 and 0.91 where few false positives were observed when a test data set was analyzed (Figure 3). This figure shows 19 minimotifs that are colored blue indicating support by experiments in the literature; we note that two of these minimotifs have scores below 0.24 and three do not have scores because of missing information. Of the other seven high scoring predicted minimotifs, two of these minimotifs were for previously known interactions of Nef with the SH3 domain of Fyn and with AIP-1; both were annotated and added to the MnM database (41,42). One was a for a c-Raf1 binding motif consensus sequence where there is a verified instance. A minimotif was identified for binding to the β subunit of AP1, AP2 and AP3, which was previously known and has now been added to MnM; the motif predicted to interact with the AP2 and AP3 µ subunit was not previously identified (43–46). MnM predicted phosphorylation of Nef by PKCα at three sites: 15, 80 and 103, none of which were present in MnM. In support of these predictions, Nef is known to be phosphorylated at Thr 15, which is inhibited by a PKC inhibitor (47,48). We note that only 29% of >7000 HIV isolate sequences have a Thr in this position of Nef; whereas 98% of these viruses have a Ser at position 103 [analysis with HIVToolbox (40)]. Ser 103 was suggested to be phosphorylated by PKCα in vitro (49). Thr at position 80 was predicted as a PKCα site, but no evidence supporting phosphorylation of this site by PKCα could be identified in the literature. MnM also predicted a novel interactions of Nef with the C-terminal SH3 domain of Grb2 and a site that binds peptidylprolyl isomerase. In summary, of the 24 minimotifs identified by the MnM analysis (including one with three distinct sites) with scores above a major false-positive threshold, 21 had previously been demonstrated and we cannot rule out the possibility that the other three have not yet been discovered. Although Nef is well studied, most proteins have many minimotifs predicted with scores above 0.24 that are yet to be investigated.
Figure 3.
Results table in MnM 3 from analysis of HIV-1 Nef protein. Nef (NP_057857) was analyzed to produce the minimotif predictions shown. Column 1 shows the minimotif sequence, column 2 shows the function of the minimotif, column 3 shows the amino acid position(s) for the start residue in the minimotif, column 4 shows the combined filter score, and column 5 shows the number of occurrences of each motif in the entire HIV-1 proteome. Rows colored blue are for minimotifs that are experimentally validated, yellow are above a threshold for high accuracy prediction, and red are below this threshold or do not have data to calculate a combined filter score (null).
Some scientists may want to analyze many protein sequences at once. We have now enabled this type of workflow as an email service for batch query input mode on the MnM input page. The input file for the request must contain a list of protein accession numbers from one or more various data sources (UniProtKB, MIM, RefSeq, Ensemble, UniGene, MIM, PIR, Entrez Gene) and/or protein sequences; this format is indicated in a hyperlink in this section of the input page.
Another workflow is identifying minimotifs that play a role in human disease, or organism diversity as originally reported (27,50). In MnM 2, this was accomplished by mapping missense SNPs located in protein coding regions from the dbSNP database (4,39). In the View menu on the results page, the ‘View SNPs’ selection reveals known SNPs in the Protein Sequence window highlighted blue and capitalized. When any SNP is clicked, the SNP has a green highlight and the amino acid change is shown. Any combination of SNPs can be selected. The ‘View motifs from New SNPs’ found under the ‘View’ menu item will create a new table that identifies any minimotifs that are introduced or eliminated by the selected SNPs. Since many SNPs are for disease-associated mutations, this tool can be used to formulate new hypotheses about disease mechanisms.
DISCUSSION AND CONCLUSIONS
We have expanded the model for Minimotifs to contain 28 attributes that offers a number of advantages. Some advantages are the segregation of specific attributes are that it reduces ambiguities and faulty annotations, can be readily used to identify missing data, and allows the use of many different types of controlled vocabularies. The rich model enables easy mining of data through SQL queries in a number of ways. For example, MnM allows a widget-based custom query builder to mine Query Engine MnM database. Using this tool, consensus sequences or position-specific scoring matrices can be generated for minimotifs where many different instances were studied, often in separate laboratories. In this manner, our model that maintains the instance information in its raw form becomes a rich source for automated generation of consensus motifs.
MnM offers a unique resource that is synergistic with other minimotif search tools. MnM is a broad-based minimotif resource that covers all types of minimotifs from any species with now approximately 300 000 minimotifs. A brief comparison with some other motifs tools is highlighted, but there are far too many tools to present a comprehensive review. The Eukaryotic Linear Motif Server is the closest broad-based minimotif resource with 170 consensus motifs and 1817 instances (6). Phospho-ELM, an associated database that focuses on phosphorylation sites has approximately 42 000 instances (5). These tools use a different, but overlapping set of approaches to help reduce false positives. Other sites such as Scansite and DomPep use position specific scoring matrices for predicting new instances, but focus on a set of protein binding domains (3,10). MOTIPs can be used to search proteomes for minimotifs (9). SLIMSearch 2.0 and MyHits allow proteome search of user-defined motifs (8,51).
Minimotif Miner 3 is an important improvement over MnM 2. The number of minimotif sequences has increased two orders of magnitude, vastly improving the sensitivity of minimotif search. This large increase in the number of potential minimotifs could potentially hinder researchers rather than help if not for the aid of filtering mechanisms to reduce the number of false positives. The new filtering mechanisms recently introduced, based on protein–protein interactions, molecular function, cellular function, genetic interactions and the combined filter, greatly improve the accuracy and specificity of MnM 3 search results.
AVAILABILITY
The MnM database can be accessed through single protein or batch queries using the MnM user interface. The entire database is not currently available for download, but the MnM investigators are open to collaborations that involve using the database.
FUNDING
National Institutes of Health (GM07689, LM010101, RR016464) and National Science Foundation (1005223, 0829916). Funding for open access charge: NIH (R01GM079689).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We would like to thank past members of the MnM group who have contributed to the construction of the MnM database and web system.
REFERENCES
- 1.Puntervoll P, Linding R, Gemund C, Chabanis-Davidson S, Mattingsdal M, Cameron S, Martin DMA, Ausiello G, Brannetti B, Costantini A, et al. ELM server: a new resource for investigating short functional sites in modular eukaryotic proteins. Nucleic Acids Res. 2003;31:3625–3630. doi: 10.1093/nar/gkg545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Davey NE, Haslam NJ, Shields DC, Edwards RJ. SLiMFinder: a web server to find novel, significantly over-represented, short protein motifs. Nucleic Acids Res. 2010;38:W534–W549. doi: 10.1093/nar/gkq440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Obenauer JC, Cantley LC, Yaffe MB. Scansite 2.0: proteome-wide prediction of cell signaling interactions using short sequence motifs. Nucleic Acids Res. 2003;31:3635–3641. doi: 10.1093/nar/gkg584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rajasekaran S, Balla S, Gradie P, Gryk MR, Kadaveru K, Kundeti V, Maciejewski MW, Mi T, Rubino N, Vyas J, et al. Minimotif miner 2nd release: a database and web system for motif search. Nucleic Acids Res. 2009;37:D185–D190. doi: 10.1093/nar/gkn865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dinkel H, Chica C, Via A, Gould CM, Jensen LJ, Gibson TJ, Diella F. Phospho.ELM: a database of phosphorylation sites–update 2011. Nucleic Acids Res. 2011;39:D261–D267. doi: 10.1093/nar/gkq1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gould CM, Diella F, Via A, Puntervoll P, Gemünd C, Chabanis-Davidson S, Michael S, Sayadi A, Bryne JC, Chica C, et al. ELM: the status of the 2010 eukaryotic linear motif resource. Nucleic Acids Res. 2010;38:D167–D180. doi: 10.1093/nar/gkp1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Balla S, Thapar V, Verma S, Luong T, Faghri T, Huang CH, Rajasekaran S, del Campo JJ, Shinn JH, Mohler WA, et al. Minimotif Miner: a tool for investigating protein function. Nat. Methods. 2006;3:175–177. doi: 10.1038/nmeth856. [DOI] [PubMed] [Google Scholar]
- 8.Davey NE, Haslam NJ, Shields DC, Edwards RJ. SLiMSearch 2.0: biological context for short linear motifs in proteins. Nucleic Acids Res. 2011;39:W56–W60. doi: 10.1093/nar/gkr402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lam HYK, Kim PM, Mok J, Tonikian R, Sidhu SS, Turk BE, Snyder M, Gerstein MB. MOTIPS: automated motif analysis for predicting targets of modular protein domains. BMC Bioinformatics. 2010;11:243. doi: 10.1186/1471-2105-11-243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li L, Zhao B, Du J, Zhang K, Ling CX, Li SS-C. DomPep-A general method for predicting modular domain-mediated protein-protein interactions. PLoS One. 2011;6:e25528. doi: 10.1371/journal.pone.0025528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rajasekaran S, Merlin JC, Kundeti V, Mi T, Oommen A, Vyas J, Alaniz I, Chung K, Chowdhury F, Deverasatty S, et al. A computational tool for identifying minimotifs in protein-protein interactions and improving the accuracy of minimotif predictions. Proteins. 2010;79:153–164. doi: 10.1002/prot.22868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vyas J, Nowling RJ, Maciejewski MW, Rajasekaran S, Gryk MR, Schiller MR. A proposed syntax for Minimotif Semantics, version 1. BMC Genomics. 2009;10:360. doi: 10.1186/1471-2164-10-360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rajasekaran S, Mi T, Merlin JC, Oommen A, Gradie P, Schiller MR. Partitioning of minimotifs based on function with improved prediction accuracy. PLoS One. 2010;5:e12276. doi: 10.1371/journal.pone.0012276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stein A, Céol A, Aloy P. 3did: identification and classification of domain-based interactions of known three-dimensional structure. Nucleic Acids Res. 2011;39:D718–D723. doi: 10.1093/nar/gkq962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vanhee P, Reumers J, Stricher F, Baeten L, Serrano L, Schymkowitz J, Rousseau F. PepX: a structural database of non-redundant protein-peptide complexes. Nucleic Acids Res. 2010;38:D545–D551. doi: 10.1093/nar/gkp893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.London N, Movshovitz-Attias D, Schueler-Furman O. The structural basis of peptide-protein binding strategies. Structure. 2010;18:188–199. doi: 10.1016/j.str.2009.11.012. [DOI] [PubMed] [Google Scholar]
- 17.Hornbeck PV, Chabra I, Kornhauser JM, Skrzypek E, Zhang B. PhosphoSite: a bioinformatics resource dedicated to physiological protein phosphorylation. Proteomics. 2004;4:1551–1561. doi: 10.1002/pmic.200300772. [DOI] [PubMed] [Google Scholar]
- 18.Ceol A, Chatr-Aryamontri A, Santonico E, Sacco R, Castagnoli L, Cesareni G. DOMINO: a database of domain-peptide interactions. Nucleic Acids Res. 2007;35:D557–D560. doi: 10.1093/nar/gkl961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rawlings ND, Barrett AJ, Bateman A. MEROPS: the peptidase database. Nucleic Acids Res. 2010;38:D227–D233. doi: 10.1093/nar/gkp971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.The Universal Protein Resource (UniProt) in 2010. Nucleic Acids Res. 2010;38:D142–D148. doi: 10.1093/nar/gkp846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Keshava Prasad TS, Goel R, Kandasamy K, Keerthikumar S, Kumar S, Mathivanan S, Telikicherla D, Raju R, Shafreen B, Venugopal A, et al. Human Protein Reference Database–2009 update. Nucleic Acids Res. 2009;37:D767–D772. doi: 10.1093/nar/gkn892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Diella F, Haslam N, Chica C, Budd A, Michael S, Brown NP, Trave G, Gibson TJ. Understanding eukaryotic linear motifs and their role in cell signaling and regulation. Front. Biosci. 2008;13:6580–6603. doi: 10.2741/3175. [DOI] [PubMed] [Google Scholar]
- 23.Lieber DS, Elemento O, Tavazoie S. Large-scale discovery and characterization of protein regulatory motifs in eukaryotes. PLoS One. 2010;5:e14444. doi: 10.1371/journal.pone.0014444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kadaveru K, Vyas J, Schiller MR. Viral infection and human disease- insights from minimotifs. Front Biosci. 2008;13:6455–6471. doi: 10.2741/3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Davey NE, Travé G, Gibson TJ. How viruses hijack cell regulation. Trends Biochem. Sci. 2011;36:159–169. doi: 10.1016/j.tibs.2010.10.002. [DOI] [PubMed] [Google Scholar]
- 26.Evans P, Dampier W, Ungar L, Tozeren A. Prediction of HIV-1 virus-host protein interactions using virus and host sequence motifs. BMC Med. Genomics. 2009;2:27. doi: 10.1186/1755-8794-2-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kadaveru K, Vyas J, Schiller MR. Viral infection and human disease - insights from minimotifs. Front. Biosci. 2008;13:6455–6471. doi: 10.2741/3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Parthasarathi L, Casey F, Stein A, Aloy P, Shields DC. Approved drug mimics of short peptide ligands from protein interaction motifs. J. Chem. Inf. Model. 2008;48:1943–1948. doi: 10.1021/ci800174c. [DOI] [PubMed] [Google Scholar]
- 29.Aasland R, Abrams C, Ampe C, Ball LJ, Bedford MT, Cesareni G, Gimona M, Hurley JH, Jarchau T, Lehto VP, et al. Normalization of nomenclature for peptide motifs as ligands of modular protein domains. FEBS Lett. 2002;513:141–144. doi: 10.1016/s0014-5793(01)03295-1. [DOI] [PubMed] [Google Scholar]
- 30.Montecchi-Palazzi L, Beavis R, Binz P-A, Chalkley RJ, Cottrell J, Creasy D, Shofstahl J, Seymour SL, Garavelli JS. The PSI-MOD community standard for representation of protein modification data. Nat. Biotechnol. 2008;26:864–866. doi: 10.1038/nbt0808-864. [DOI] [PubMed] [Google Scholar]
- 31.Vyas J, Nowling RJ, Meusburger T, Sargeant D, Kadaveru K, Gryk MR, Kundeti V, Rajasekaran S, Schiller MR. MimoSA: a system for minimotif annotation. BMC Bioinformatics. 2010;11 doi: 10.1186/1471-2105-11-328. 328, doi: 10.1186/1471-2105-11-328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gould CM, Diella F, Via A, Puntervoll P, Gemünd C, Chabanis-Davidson S, Michael S, Sayadi A, Bryne JC, Chica C, et al. ELM: the status of the 2010 eukaryotic linear motif resource. Nucleic Acids Res. 2010;38:D167–D180. doi: 10.1093/nar/gkp1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chatr-Aryamontri A, Ceol A, Peluso D, Nardozza A, Panni S, Sacco F, Tinti M, Smolyar A, Castagnoli L, Vidal M, et al. VirusMINT: a viral protein interaction database. Nucleic Acids Res. 2009;37:D669–D673. doi: 10.1093/nar/gkn739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ceol A, Chatr Aryamontri A, Licata L, Peluso D, Briganti L, Perfetto L, Castagnoli L, Cesareni G. MINT, the molecular interaction database: 2009 update. Nucleic Acids Res. 2010;38:D532–D539. doi: 10.1093/nar/gkp983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xenarios I, Salwínski L, Duan XJ, Higney P, Kim S-M, Eisenberg D. DIP, the Database of Interacting Proteins: a research tool for studying cellular networks of protein interactions. Nucleic Acids Res. 2002;30:303–305. doi: 10.1093/nar/30.1.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sayers EW, Barrett T, Benson DA, Bolton E, Bryant SH, Canese K, Chetvernin V, Church DM, Dicuccio M, Federhen S, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2010;38:D5–D16. doi: 10.1093/nar/gkp967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Aranda B, Achuthan P, Alam-Faruque Y, Armean I, Bridge A, Derow C, Feuermann M, Ghanbarian AT, Kerrien S, Khadake J, et al. The IntAct molecular interaction database in 2010. Nucleic Acids Res. 2010;38:D525–D531. doi: 10.1093/nar/gkp878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.The Gene Ontology Consortium. The Gene Ontology in 2010: extensions and refinements. Nucleic Acids Res. 2010;38:D331–D335. doi: 10.1093/nar/gkp1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sayers EW, Barrett T, Benson DA, Bolton E, Bryant SH, Canese K, Chetvernin V, Church DM, Dicuccio M, Federhen S, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2010;38:D5–D16. doi: 10.1093/nar/gkp967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sargeant D, Deverasatty S, Luo Y, Baleta AV, Zobrist S, Rathnayake V, Russo JC, Vyas J, Muesing MA, Schiller MR. HIVToolbox, an integrated web application for investigating HIV. PloS One. 2011;6:e20122. doi: 10.1371/journal.pone.0020122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Costa LJ, Chen N, Lopes A, Aguiar RS, Tanuri A, Plemenitas A, Peterlin BM. Interactions between Nef and AIP1 proliferate multivesicular bodies and facilitate egress of HIV-1. Retrovirology. 2006;3:33. doi: 10.1186/1742-4690-3-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Arold S, Franken P, Strub MP, Hoh F, Benichou S, Benarous R, Dumas C. The crystal structure of HIV-1 Nef protein bound to the Fyn kinase SH3 domain suggests a role for this complex in altered T cell receptor signaling. Structure. 1997;5:1361–1372. doi: 10.1016/s0969-2126(97)00286-4. [DOI] [PubMed] [Google Scholar]
- 43.Lindwasser OW, Smith WJ, Chaudhuri R, Yang P, Hurley JH, Bonifacino JS. A diacidic motif in human immunodeficiency virus type 1 Nef is a novel determinant of binding to AP-2. J. Virol. 2008;82:1166–1174. doi: 10.1128/JVI.01874-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Coleman SH, Madrid R, Van Damme N, Mitchell RS, Bouchet J, Servant C, Pillai S, Benichou S, Guatelli JC. Modulation of cellular protein trafficking by human immunodeficiency virus type 1 Nef: role of the acidic residue in the ExxxLL motif. J. Virol. 2006;80:1837–1849. doi: 10.1128/JVI.80.4.1837-1849.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Greenberg M, DeTulleo L, Rapoport I, Skowronski J, Kirchhausen T. A dileucine motif in HIV-1 Nef is essential for sorting into clathrin-coated pits and for downregulation of CD4. Curr. Biol. 1998;8:1239–1242. doi: 10.1016/s0960-9822(07)00518-0. [DOI] [PubMed] [Google Scholar]
- 46.Coleman SH, Van Damme N, Day JR, Noviello CM, Hitchin D, Madrid R, Benichou S, Guatelli JC. Leucine-specific, functional interactions between human immunodeficiency virus type 1 Nef and adaptor protein complexes. J. Virol. 2005;79:2066–2078. doi: 10.1128/JVI.79.4.2066-2078.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bandres JC, Luria S, Ratner L. Regulation of human immunodeficiency virus Nef protein by phosphorylation. Virology. 1994;201:157–161. doi: 10.1006/viro.1994.1278. [DOI] [PubMed] [Google Scholar]
- 48.Bodéus M, Marie-Cardine A, Bougeret C, Ramos-Morales F, Benarous R. In vitro binding and phosphorylation of human immunodeficiency virus type 1 Nef protein by serine/threonine protein kinase. J. Gen. Virol. 1995;76(Pt 6):1337–1344. doi: 10.1099/0022-1317-76-6-1337. [DOI] [PubMed] [Google Scholar]
- 49.Coates K, Harris M. The human immunodeficiency virus type 1 Nef protein functions as a protein kinase C substrate in vitro. J. Gen. Virol. 1995;76(Pt 4):837–844. doi: 10.1099/0022-1317-76-4-837. [DOI] [PubMed] [Google Scholar]
- 50.Schiller MR. Minimotif Miner: a computation tool to investigate protein function, disease, and genetic diversity. In: Coligan JE, Dunn BM, Speicher DW, Winkler H, editors. Current Protocols in Protein Science. New York: John Wiley & Sons, Inc.; 2007. pp. 2.12.1–2.12.14. [DOI] [PubMed] [Google Scholar]
- 51.Pagni M, Ioannidis V, Cerutti L, Zahn-Zabal M, Jongeneel CV, Hau J, Martin O, Kuznetsov D, Falquet L. MyHits: improvements to an interactive resource for analyzing protein sequences. Nucleic Acids Res. 2007;35:W433–W437. doi: 10.1093/nar/gkm352. [DOI] [PMC free article] [PubMed] [Google Scholar]