Abstract
The Department of Energy (DOE) Joint Genome Institute (JGI) is a national user facility with massive-scale DNA sequencing and analysis capabilities dedicated to advancing genomics for bioenergy and environmental applications. Beyond generating tens of trillions of DNA bases annually, the Institute develops and maintains data management systems and specialized analytical capabilities to manage and interpret complex genomic data sets, and to enable an expanding community of users around the world to analyze these data in different contexts over the web. The JGI Genome Portal (http://genome.jgi.doe.gov) provides a unified access point to all JGI genomic databases and analytical tools. A user can find all DOE JGI sequencing projects and their status, search for and download assemblies and annotations of sequenced genomes, and interactively explore those genomes and compare them with other sequenced microbes, fungi, plants or metagenomes using specialized systems tailored to each particular class of organisms. We describe here the general organization of the Genome Portal and the most recent addition, MycoCosm (http://jgi.doe.gov/fungi), a new integrated fungal genomics resource.
INTRODUCTION
Established in 1997, the DOE JGI united the expertise and resources in DNA sequencing, informatics and technology development pioneered at three national laboratories to work on the Human Genome Project. Seven years later, the DOE JGI became a national user facility targeting research relevant to the DOE mission areas of bioenergy, carbon cycling and biogeochemistry. The DOE JGI leads the world in the number of organisms sequenced in four areas: plants, fungi, microbes and metagenomes [according to GOLD: Genomes Online Database (1)].
Aside from generating and storing sequence, the Institute has developed a wide array of databases and analytical systems to interpret the data. Some systems work across multiple JGI databases, while others allow users to specifically manage data sets on plants (Phytozome) (D. M. Goodstein et al. submitted for publication), fungi (MycoCosm, described here), microbes (Integrated Microbial Genomes or IMG) (2) and both metagenomes and single cells (IMG/M) (3). In addition to plants and fungi, diverse eukaryotes from Amoebozoa (4), Metazoa (5), Choanozoa (6), Heterobosea (7), Heterokonta (8–10), Rhizaria, Haptophyta and Cryptophyta can be analyzed with a collection of tools linked directly to their genome databases.
The Genome Portal (http://genome.jgi.doe.gov) provides a unified access point and navigation capabilities for multiple interconnected resources, both for general and specialized use. Different stages of a genome project require different tools for data access and analysis. Here, we walk through JGI systems for data access and analysis at three major stages of genome projects: tracking projects, getting access to genome sequences and annotations, and interactively exploring genomic data. Building specialized tools for efficient analysis and exploration of the constantly growing number of genomes is critically important. MycoCosm (http://jgi.doe.gov/fungi), first released in 2010, provides access to the database of over a hundred of fungal genomes and a number of analytical tools for the DOE JGI Fungal Genomics program.
DATABASES ACCESSIBLE THROUGH INTEGRATED GENOME PORTAL
DOE JGI sequencing projects, ongoing and completed
Close to 4000 DOE JGI projects of different types are publicly available and searchable in our database. These projects include different genomic products, such as standard and improved draft, finished genomes, gene expression profiling, resequencing, metagenome projects and others. The ‘Project List’ links on the Genome Portal page (http://genome.jgi.doe.gov) and most of the Portal pages brings users to a list of DOE JGI projects with a detailed description of each project including its scope and current status, taxon, the JGI program and the project lead. The Resources column lists tools available for this project. Some of these tools, e.g. download are available for all genomes, while others are taxon-, project type- or stage-dependent. For example, the plant genome will be linked to Phytozome, and the fungal genome—to MycoCosm.
Annotated DOE JGI genomes
The Genome Portal provides unified access to all annotated genomes and metagenomes available at the DOE JGI along with specialized analytical tools to navigate these data sets and compare genomes of related organisms. It is available at http://genome.jgi.doe.gov or via the ‘Genomes’ tab on the JGI home page http://www.jgi.doe.gov/. The Portal home page also provides worldwide statistics on the usage of the JGI resources and the information about the latest genome releases and new tool development.
From this page a user selects the organism and/or the tools to work with. There are over 3500 annotated genomes in the JGI database, and three convenient ways to find a particular genome of interest: an interactive The Tree of Life, a selection menu on the top of the page, and the Search function.
The Tree of Life organizes the sequenced genomes by domains of life and links to Organism home pages. Clicking on a branch name produces a menu displaying available genomes in this kingdom, phylum, class, or order (Figure 1). Selecting a genome connects a user to a corresponding organism page or pages in different resources.
Figure 1.
The Genome Portal page. A pull-down menu for the ‘Fungi’ branch of Eukaryota is shown. Search, BLAST and Download functions are available for the entire selected group. Each genome is linked to the organism page in the related resources, such as Mycocosm and IMG. ‘Project list’ on the top leads users to the list of all sequencing projects at the DOE JGI. The bottom portion of the page connects to the specialized databases for microbes (IMG) and metagenomes (IMG/M), fungi (MycoCosm) and plants (Phytozome).
The same result can be achieved using the selection menu on the top of the page that allows for step-by-step genome selection by choosing All JGI Genomes, Bacteria, Archaea, Eukaryotic or Metagenome first, then organisms available for this group and finally the page to view. The latest addition to the JGI Genome Portal is Search function that enables searching for genomes by keyword (e.g. plants, Eukaryota), name, taxonId or projectId. Typing the beginning of the word in the text window brings up a pull-down menu with relevant search term choices.
Each organism's home page contains a description of the project, BLAST, download and links to specialized resources. For many eukaryotes (5–11) the menu also includes several analytical tools described in the next section. The specialized JGI database resources connected to the portal include Integrated Microbial Genomes (IMG) (2) and Metagenomes (IMG/M) (3); Phytozome for green plant genomes (D. M. Goodstein et al., submitted for publication) and MycoCosm—the Fungal Genomics Resource that provides access to the annotated fungal genomes and tools for their analysis as described further in the text.
MycoCosm, an integrated fungal genomics resource
MycoCosm (http://jgi.doe.gov/fungi) was released in March 2010, in response to a call from the fungal community for integration of all fungal genomes and analytical tools in one place. MycoCosm brings together fungal genomics data and interactive analytical tools for diverse fungi that are important for energy and environment, which is the focus of the JGI Fungal program (12,13). MycoCosm integrates genomics data from the DOE JGI and its users and promotes user community participation in data submission, annotation and analysis.
Over 100 newly sequenced and annotated fungal genomes from JGI and elsewhere are available to the public through MycoCosm, and new annotated genomes are being added to this resource upon completion of annotation. MycoCosm offers web-based genome analysis tools for fungal biologists to ‘navigate’ through sequenced genomes and explore them in the context of ‘genome-centric’ and ‘comparative views’.
MycoCosm Navigator
provides search capabilities for annotated fungal genomes and visual navigation across their phylogenetic tree, where each node represents a group of phylogenetically related organisms and links to both genome centric and comparative analysis tools (Figure 2). Each node includes a list of organisms and enables search and analysis within this list. Thus, by clicking on different nodes of the tree, a user can adjust the search and analysis space from single organism to the entire list of fungi. The Search function allows users to type an organism name or part of it and jump directly to a specific genome without browsing the tree.
Figure 2.
The MycoCosm home page includes genome search function and displays major branches of the Fungal Tree of Life with nodes representing phylogenetically related groups. Clicking on a node brings up drop-down menu (shown on lower right) linked to an integrated comparative view (e.g. Mucoromycotina), individual comparative tools (search, BLAST, download) and the list of sequenced genomes from this group, each linked to its own genome-centric view.
MycoCosm genome-centric view
Includes the genome browser, download, BLAST and search capabilities within the data for a single genome, the VISTA tools for the analysis of whole-genome alignments, functional profiles and gene clusters (Figure 3).
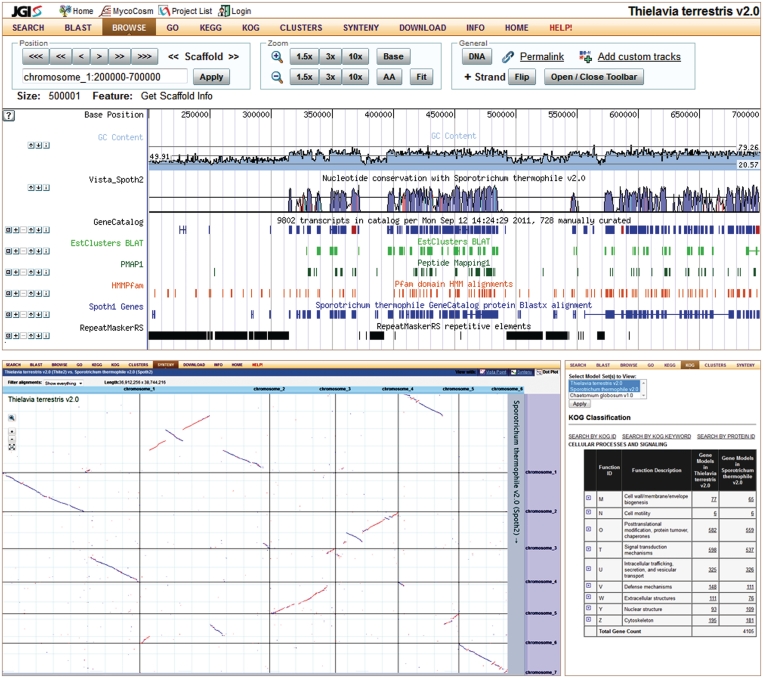
Figure 3.
Genome-centric view of the MycoCosm includes several tools (listed in the top menu) and illustrated here by Genome Browser (on top), Synteny by interactive VISTADot plot (lower left) and KOG functional profile (lower right). Genome Browser tracks shown for a thermophile Thielavia terrestris (14) include GC content (light blue), VISTA-based genome conservation (blue and red curve), automatically predicted (blue) and manually curated (red) gene models, transcriptomics (light green) and proteomics (dark green) data, PFAM domains (orange), BLASTx hits against proteins of related organism (blue), and repeats (black). Dot plot is based on VISTA whole genome alignments of two genomes and interactively displays syntenic blocks (collinear in blue or anti-sense in red). KOG profile summarizes functional annotations of genes according to this classification and allows comparison of gene counts in each category between related genomes (last two columns).
The Genome browser
is the centerpiece of the MycoCosm genome-centric view and is based on the earlier version of the UCSC Genome Browser (15) with configurable selection of tracks (Figure 3). It displays predicted gene models and annotations along with different lines of evidence in support of these predictions (e.g. gene and protein expression profiles). It also displays other types of data mapped to a genome assembly such as VISTA tracks of genome conservation (16), G+C profiles and annotation features including regions of homology, domains, repeats, non-coding genes and others. These features are either automatically computed or loaded by registered users as custom tracks. Predicted genome features in each track are linked to pages describing them and can also be linked to external resources. Gene models tracks are linked to the annotation reports and community annotation tools, which allow registered users to revise the predicted annotations.
Community annotation
This is a unique model across sequencing centers developed by the DOE JGI to engage users in collective analysis and improvement of genome annotations, which resulted in many successful projects (14,17–19). Registered users participating in a particular genome project can validate and improve predicted gene models and annotations. Such gene models become highlighted on the browser (Figure 3). Structural modifications are supported by the tools linked to Genome browser, which allow users to copy exons and gene models from any track, change them, or create them de novo. Functional annotation tools are linked to annotation reports and enable user to curate functional assignments such as gene name and description, and communicate with other annotators.
Functional profiles of genomes are based on summaries of predicted gene annotations according to the GO (20), KEGG (21) and KOG (22) classifications. Each profile is accessible as a separate tab and is searchable according to the classification nomenclature (Figure 3). The profile lists the numbers of genes assigned to a particular functional category in the classification and links each number to the list of proteins assigned to the category. For every reference genome, a user can also compare its functional profile with profiles of related genomes to investigate gene family expansions or contractions at different levels of granularity.
Genome conservation and synteny can be explored using VISTA Point, designed for visualization and analysis of pairwise- and multiple DNA alignments (16) at different levels of resolution in three visualization modes: (i) VISTA Browser, which enables visual comparative analysis of complete genome assemblies using pairwise and multiple large-scale alignments; (ii) VISTA Synteny Viewer, a multi-tiered graphical display of pairwise alignments at three different levels of resolution; (iii) VistaDot, an interactive two-dimensional dot-plot genome synteny viewer across multiple chromosomes/scaffolds (Figure 3). VISTA tools are also available through Phytozome and IMG for the plant and microbial genomes, respectively.
MycoCosm comparative view
This provides a different context for analyzing and summarizing information for entire groups of genomes, predefined in MycoCosm and corresponding to its nodes (Figure 2). Unlike the genome-centric view, there is no reference genome in this analysis. Therefore, BLAST and search functions in this view are distinct from the genome-centric versions by their ability to search across multiple genomes simultaneously and compare analysis results side by side. For example, a keyword or BLAST search for protein kinases in Basidiomycota or Ascomycota will show differences in the number of found genes or BLAST hits across different members of these phyla. In addition, a user can save and download search results in different formats (FASTA, GFF) or download sequences and annotations for an entire group of organisms or its subset using the download tab.
Clusters analysis
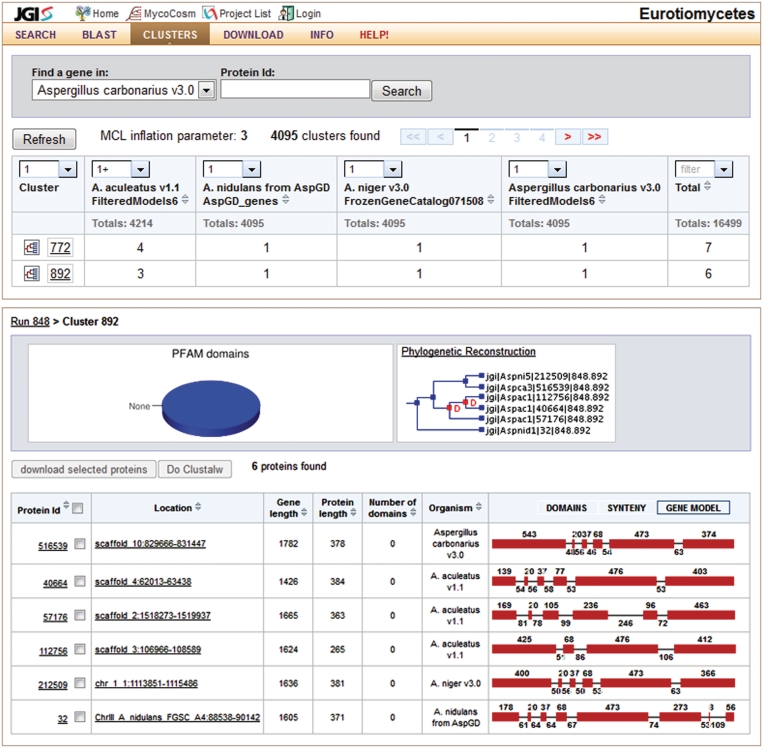
This enables exploration of gene families within a given group of organisms. Clusters are built using Markov clustering algorithm MCL (23) and all-against-all BLAST alignments of the proteins from the entire data set. On the Clusters front page, a user will find clusters of interest using gene search or cardinality filters to identify genome-specific clusters or those conserved across multiple genomes from the group (Figure 4). Each cluster is linked to the Cluster Details page, where a user can explore the pattern of protein domains, intron–exon structure and local genomic context of each of the cluster members side-by-side. For some clusters a user can also examine precomputed multiple alignment of protein sequences and a species-reconciled phylogenetic tree with predicted gain/loss of genes.
Figure 4.
MycoCosm comparative view includes several functions designed for analyzing groups of genomes (listed in the top menu) as illustrated by Cluster view. The Cluster front page (on top) lists two largest clusters of genes conserved in all four Eurotiomycetes and expanded in Aspergillus aculeatus, after using filters (1+:1:1:1). Cluster details page (on bottom) shows six members of the cluster 892, their intron–exon gene structures (right column), PFAM domain composition (pie chart in the middle, no predicted domains here) and species-reconciliated gene tree suggesting two gene duplications Aspergillus carbonarius (red nodes D on the tree in the middle).
On-line video tutorial
This is available from the link on the main MycoCosm page (Figure 2). It provides additional information on all features of MycoCosm and walks a user through the genome analysis process step by step. Several analytical tools are also available outside of MycoCosm for other eukaryotes (4–11).
Architecture
The Genome Portal web site is built on Apache HTTPD, Tomcat and MySQL. A majority of the Genome Portal components has been developed using Java and a variety of available open-sources tools and technologies. Our scalable database architecture is based on MySQL servers and currently contains more than 25 TB of genomics data. There are four load-balanced web servers, talking to two back-end database servers. A web-driven automated build system that takes each machine silently out of the cluster, builds a new version of the portal and puts the machine back into the cluster, ensures that updates can be applied without disruption to users. This setup further makes the portal resilient against hardware failures.
Data is fed into the portal by the JGI's annotation pipelines via an API that makes the data available to authorized users immediately. An advanced monitoring system allows administrators to quickly assess issues and deal with them before they become problems that may impact web site and database performance.
FUTURE PLANS
Democratization of genome sequencing, and the low cost and high quantities of data being produced by new sequencing technologies will result in avalanche of new sequenced genomes. The DOE JGI Fungal Genomics program alone aims to double sequencing and analysis throughput every year. This requires new analytical tools, further scalability in data storage and better integration for the DOE JGI to continue to enable science and serve as a central hub for user communities.
FUNDING
Director, Office of Science, Office of Biological and Environmental Research, Life Sciences Division, U.S. Department of Energy (Contract No. DE-AC02-05CH11231). Funding for open access charge: DOE JGI.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We would like to thank the large team of people who worked on the development of the JGI Genome Portal through the long history of this project: Paramvir Dehal, Serge Dusheyko, Kelly Felkins, Martin Gelkpe, Annette Greiner, Leila Hornick, Katherine Huang, Wayne Huang, Jinal Jhaveri, Sam Rash, Lukasz Szajkowski, Gregory Werner and the JGI user community for using these tools and providing valuable feedback. We are thankful to David Gilbert for his help with the manuscript. Eddy Rubin, James Bristow and Victor Markowitz provided support, advice and encouragement throughout this project.
REFERENCES
- 1.Liolios K, Chen IM, Mavromatis K, Tavernarakis N, Hugenholtz P, Markowitz VM, Kyrpides NC. The Genomes On Line Database (GOLD) in 2009: status of genomic and metagenomic projects and their associated metadata. Nucleic Acids Res. 2010;38:D346–D354. doi: 10.1093/nar/gkp848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Markowitz VM, Szeto E, Palaniappan K, Grechkin Y, Chu K, Chen IM, Dubchak I, Anderson I, Lykidis A, Mavromatis K, et al. The integrated microbial genomes (IMG) system in 2007: data content and analysis tool extensions. Nucleic Acids Res. 2008;36:D528–D533. doi: 10.1093/nar/gkm846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Markowitz VM, Ivanova NN, Szeto E, Palaniappan K, Chu K, Dalevi D, Chen IM, Grechkin Y, Dubchak I, Anderson I, et al. IMG/M: a data management and analysis system for metagenomes. Nucleic Acids Res. 2008;36:D534–D538. doi: 10.1093/nar/gkm869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sucgang R, Kuo A, Tian X, Salerno W, Parikh A, Feasley CL, Dalin E, Tu H, Huang E, Barry K, et al. Comparative genomics of the social amoebae Dictyostelium discoideum and Dictyostelium purpureum. Genome Biol. 2011;12:R20. doi: 10.1186/gb-2011-12-2-r20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Colbourne JK, Pfrender ME, Gilbert D, Thomas WK, Tucker A, Oakley TH, Tokishita S, Aerts A, Arnold GJ, Basu MK, et al. The ecoresponsive genome of Daphnia pulex. Science. 2011;331:555–561. doi: 10.1126/science.1197761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.King N, Westbrook MJ, Young SL, Kuo A, Abedin M, Chapman J, Fairclough S, Hellsten U, Isogai Y, Letunic I, et al. The genome of the choanoflagellate Monosiga brevicollis and the origin of metazoans. Nature. 2008;451:783–788. doi: 10.1038/nature06617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fritz-Laylin LK, Prochnik SE, Ginger ML, Dacks JB, Carpenter ML, Field MC, Kuo A, Paredez A, Chapman J, Pham J, et al. The genome of Naegleria gruberi illuminates early eukaryotic versatility. Cell. 2010;140:631–642. doi: 10.1016/j.cell.2010.01.032. [DOI] [PubMed] [Google Scholar]
- 8.Gobler CJ, Berry DL, Dyhrman ST, Wilhelm SW, Salamov A, Lobanov AV, Zhang Y, Collier JL, Wurch LL, Kustka AB, et al. Niche of harmful alga Aureococcus anophagefferens revealed through ecogenomics. Proc. Natl Acad. Sci. USA. 2011;108:4352–4357. doi: 10.1073/pnas.1016106108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bowler C, Allen AE, Badger JH, Grimwood J, Jabbari K, Kuo A, Maheswari U, Martens C, Maumus F, Otillar RP, et al. The Phaeodactylum genome reveals the evolutionary history of diatom genomes. Nature. 2008;456:239–244. doi: 10.1038/nature07410. [DOI] [PubMed] [Google Scholar]
- 10.Tyler BM, Tripathy S, Zhang X, Dehal P, Jiang RH, Aerts A, Arredondo FD, Baxter L, Bensasson D, Beynon JL, et al. Phytophthora genome sequences uncover evolutionary origins and mechanisms of pathogenesis. Science. 2006;313:1261–1266. doi: 10.1126/science.1128796. [DOI] [PubMed] [Google Scholar]
- 11.Worden AZ, Lee JH, Mock T, Rouze P, Simmons MP, Aerts AL, Allen AE, Cuvelier ML, Derelle E, Everett MV, et al. Green evolution and dynamic adaptations revealed by genomes of the marine picoeukaryotes Micromonas. Science. 2009;324:268–272. doi: 10.1126/science.1167222. [DOI] [PubMed] [Google Scholar]
- 12.Grigoriev IV, Cullen D, Goodwin SB, Hibbett D, Jeffries TW, Kubicek CP, Kuske C, Magnuson JK, Martin F, Spatafora JW, et al. Fueling the future with fungal genomics. Mycology. 2011 (doi:10.1080/21501203.2011.584577; epub ahead of print) [Google Scholar]
- 13.Martin F, Cullen D, Hibbett D, Pisabarro A, Spatafora JW, Baker SE, Grigoriev IV. Sequencing the fungal tree of life. New Phytol. 2011;190:818–821. doi: 10.1111/j.1469-8137.2011.03688.x. [DOI] [PubMed] [Google Scholar]
- 14.Berka RM, Grigoriev IV, Otillar R, Salamov A, Grimwood J, Reid I, Ishmael N, John T, Darmond C, Moisan MC, et al. Comparative genomic analysis of the thermophilic biomass-degrading fungi Myceliophthora thermophila and Thielavia terrestris. Nat. Biotechnol. 2011 doi: 10.1038/nbt.1976. [DOI] [PubMed] [Google Scholar]
- 15.Fujita PA, Rhead B, Zweig AS, Hinrichs AS, Karolchik D, Cline MS, Goldman M, Barber GP, Clawson H, Coelho A, et al. The UCSC Genome Browser database: update 2011. Nucleic Acids Res. 2011;39:D876–D882. doi: 10.1093/nar/gkq963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Frazer KA, Pachter L, Poliakov A, Rubin EM, Dubchak I. VISTA: computational tools for comparative genomics. Nucleic Acids Res. 2004;32:W273–W279. doi: 10.1093/nar/gkh458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eastwood DC, Floudas D, Binder M, Majcherczyk A, Schneider P, Aerts A, Asiegbu FO, Baker SE, Barry K, Bendiksby M, et al. The plant cell wall-decomposing machinery underlies the functional diversity of forest fungi. Science. 2011;333:762–765. doi: 10.1126/science.1205411. [DOI] [PubMed] [Google Scholar]
- 18.Martin F, Aerts A, Ahren D, Brun A, Danchin EG, Duchaussoy F, Gibon J, Kohler A, Lindquist E, Pereda V, et al. The genome of Laccaria bicolor provides insights into mycorrhizal symbiosis. Nature. 2008;452:88–92. doi: 10.1038/nature06556. [DOI] [PubMed] [Google Scholar]
- 19.Ohm RA, de Jong JF, Lugones LG, Aerts A, Kothe E, Stajich JE, de Vries RP, Record E, Levasseur A, Baker SE, et al. Genome sequence of the model mushroom Schizophyllum commune. Nat. Biotechnol. 2010;28:957–963. doi: 10.1038/nbt.1643. [DOI] [PubMed] [Google Scholar]
- 20.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kanehisa M, Araki M, Goto S, Hattori M, Hirakawa M, Itoh M, Katayama T, Kawashima S, Okuda S, Tokimatsu T, et al. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008;36:D480–D484. doi: 10.1093/nar/gkm882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Koonin EV, Fedorova ND, Jackson JD, Jacobs AR, Krylov DM, Makarova KS, Mazumder R, Mekhedov SL, Nikolskaya AN, Rao BS, et al. A comprehensive evolutionary classification of proteins encoded in complete eukaryotic genomes. Genome Biol. 2004;5:R7. doi: 10.1186/gb-2004-5-2-r7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Enright AJ, Van Dongen S, Ouzounis CA. An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res. 2002;30:1575–1584. doi: 10.1093/nar/30.7.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]