Abstract
Notophthalmus viridescens, a member of the salamander family is an excellent model organism to study regenerative processes due to its unique ability to replace lost appendages and to repair internal organs. Molecular insights into regenerative events have been severely hampered by the lack of genomic, transcriptomic and proteomic data, as well as an appropriate database to store such novel information. Here, we describe ‘Newt-omics’ (http://newt-omics.mpi-bn.mpg.de), a database, which enables researchers to locate, retrieve and store data sets dedicated to the molecular characterization of newts. Newt-omics is a transcript-centred database, based on an Expressed Sequence Tag (EST) data set from the newt, covering ∼50 000 Sanger sequenced transcripts and a set of high-density microarray data, generated from regenerating hearts. Newt-omics also contains a large set of peptides identified by mass spectrometry, which was used to validate 13 810 ESTs as true protein coding. Newt-omics is open to implement additional high-throughput data sets without changing the database structure. Via a user-friendly interface Newt-omics allows access to a huge set of molecular data without the need for prior bioinformatical expertise.
INTRODUCTION
Comprehensive repositories for the standard model organisms mouse, rat, zebrafish, Drosophila and Xenopus (1–5) provide access to all levels of sequence data sets, including genome, transcriptome and proteome data. Detailed information for single genes usually comprises knowledge about intron and exon regions, splicing signals, as well as sequence and functional annotations. To enable a user-friendly handling of such information databases are most often accessible through graphical user interfaces that allow combinatorial searches for different properties of single genes or gene families. However, for non-standard model organisms, very little information from publically accessible data have been collected and organized in a user-friendly form. This situation prevents dissemination of useful research information to a broader research community and keeps such model organisms in splendid isolation. One of these organisms is the red spotted newt Notophthalmus viridescens (vertebrata, order caudata, family Salamandridae, genus Notophthalmus), known for its exceptional regenerative capabilities for more than 200 years. The newt does not only possess the ability to entirely replace lost appendages (6–8) but also regenerates the lens (9,10), parts of the central nervous system (11,12), and the heart (13–15). These unique features make the newt an excellent model to study fundamental processes of tissue regeneration. Newts have been helpful to study embryonic development of vertebrates (16), which led to the first Nobel prize in developmental biology. However, the introduction of other model organisms with shorter life cycles and better characterized genomes have led to a massive decline in the use of newts for basic scientific research. Moreover, the estimated genome size of the newt is up to 10 times larger than that of humans. These circumstances have severely impeded genome projects despite the increasing speed and capacities of modern sequencing machines and assembly algorithms and slowed down development of methods devoted to genetic manipulation of this animal. As a result of these drawbacks, approximately 100 non-redundant protein sequences for the newt are available in the NCBInr database, although a set of almost 11 000 sequenced Expressed Sequence Tags (ESTs) from regenerating hearts of the newt Notophthalmus viridescens exists (17).
Here, we introduce Newt-omics, the first public available data repository for N. viridescens. In addition to EST data sets and a set of high-density microarray data, Newt-omics contains a large set of peptides identified by mass spectrometry (18). Newt-omics provides a user-friendly gateway to large molecular data sets, which helps to make this attractive model organism more accessible even for researchers with limited experience in bioinformatics.
RESULTS
Transcriptome data
To expand our knowledge of transcriptional changes that occur during regeneration and to provide a data set for the Newt-omics database, we generated a normalized cDNA library from various time points after newt heart injury covering the entire cardiac regeneration process. The cDNA library was plated, 100 000 individual clones were picked, amplified and the products spotted to produce custom-made cDNA microarrays.
Complementary DNA samples from nine different time points of regenerating newt hearts plus sham controls were compared with undamaged samples. Biological and technical replicates were pre-processed and normalized. Resulting expression values for each cDNA spot were stored in the Newt-omics database along with statistical significance values and linked to their dedicated transcript. Around 52 000 EST clones were selected for Sanger sequencing based on expression changes that were detected during microarray expression analyses. After filtering, clipping and removal of contaminants, sequences were assembled into 26 594 unique transcripts with an average length of 642 bp.
Annotation of sequence data
BLAST homology searches were performed using several BLAST algorithms and databases. For this purpose, an automated annotation and quality filter pipeline was programmed. The BLAST was performed by BLASTcl3 controlled by a Unix shell script. We used blastn, blastx and tblastx on NCBI's NR (protein), NR (Nucleotide), EST and High Troughput Genomic Sequences (HTGS) databases. All hits were sorted according to their taxa. For each taxon, we performed a quality rating and filtering by regular expressions and checked for keywords providing a robust quality level for a single hit (like ‘mRNA’,‘cDNA’, ‘clone’ or ‘genomic’). The number of blast hits and their quality were calculated for each taxon independently. In total, we evaluated 90 different taxa. Since we wanted to achieve a maximum quality of annotations per transcript with a minimum number of annotations per taxon, we assumed that an annotation with a NCBI NR entry is of higher quality than an annotation with an EST entry, which by itself is of higher quality than an annotation by a HTGS entry. Based on this assumption we separated BLAST hits with the highest annotation quality from lower quality hits by computing a quality rank. The rank algorithm was based on a dictionary of keywords (e.g. cDNA, clone, mRNA, predicted) reflecting hits of limited information. Database entries containing one or more such keywords were marked as low quality hits. Our annotation was complemented by hits from lower quality groups according to the number of low quality keywords and the homology score if the minimum number of annotations was not reached for a single taxon within the highly ranked group. Our annotation process collected at least three top hits per taxon, BLAST algorithm and database. Due to this workflow, the number of selected Blast hits that were required to annotate a transcript sufficiently decreased with the extent of sequence similarities found. Interestingly, we detected a substantial number of sequenced and assembled transcripts, which did not share any reliable sequence similarity to database entries. We concluded that these sequences were either unique for urodele amphibians or had not been identified in other species yet.
Functional annotation
Subsequent to transcript annotation, we assigned transcripts to different functional groups. Since the extent of Gene Ontology (GO) annotations for amphibians is limited (19) we performed separate BLAST searches against Uniprot databases (e-value threshold < e-20) from mouse, human, zebrafish, chicken and cow. The mammalian organisms human, mouse and cow have well annotated sequence information. The chicken is the closest relative with a substantial number of GO annotated proteins in the evolutionary tree to newts. The zebrafish served as a model organism for tissue regeneration, although the annotation is comparatively poor. We only took the best-rated similarity hit per taxon with at least one GO annotation to avoid redundant assignments. GO annotations for each transcript were stored in a taxon-dependent manner in the Newt-omics database. We also assigned known functional domains, interaction partners, protein families and pathways to individual transcripts based on Uniprot entries.
Proteome data
To obtain a comprehensive set of proteins that are present during organ regeneration, we performed mass spectrometry (reverse-phase nano-LC-MS/MS) experiments on several newt heart and tail samples isolated at different time points of regeneration. We generated stable isotope labelling with amino acids in cell culture (SILAC) labelled proteins in vivo (20) to facilitate quantification and to compare protein levels between different biological samples with pulsed SILAC (21). Annotated ESTs from the transcriptome analysis were not used directly for mascot searches due to a number of inherent restrictions [discussed in detail in Ref. (18)]. Instead, all newt ESTs were reverse translated into three reading frames to generate a MASCOT-conform search database. Linkage of peptides to ESTs was achieved by computing the absolute position of the peptide within a corresponding EST. The maximum false discovery rate was set <1% for peptide and protein identifications using decoy target databases (22). We could experimentally validate 15 169 different peptides corresponding to 13 810 ESTs that were stored in the Newt-omics database.
Design of the database
The principal aim of the Newt-omics database is to provide a comprehensive and dynamic repository for ‘omics’ data from the newt N. viridescens. We intended to integrate different sets of data obtained from sequencing approaches, quantitative transcriptome or proteome studies, as well as bioinformatics analysis. The Newt-omics design is based on an Entity Relationship Model (ERM) (23) that connects data sets by biological context and avoids compartmentalization. Hence, we prioritized the selection of biological entities (such as EST, transcript and peptides) over experimental entities (such as regeneration stages of the heart). Experimental settings were generated as attributes of single entities, that allowed direct linkage between biological entities independent of their experimental origin. Entities such as EST, transcript and peptide were defined as core tables and additional entities such as transcript annotation, functional annotation of transcripts and ORF of transcripts as section tables. The central object of our database design is the transcript. Transcripts mediate queries across all sections of the database, which is the prerequisite to incorporate an advanced Graphical User Interface (GUI) search engine. A transcript table connects the main sections annotation, functional annotation, expression and peptide (Supplementary Figure S1). The database can be upgraded by implementation of future experiments. For example, new sequencing data from next generation sequencing approaches can be included into the database by adding single sequences to the EST table and by adding results of an assembly as transcripts to the transcript table. Likewise, corresponding annotations can be appended to the annotation tables. The annotation section instantiates a defined transcript and represents all annotation results of a homology search at a specific date. Furthermore, identifications of new peptides can be directly mapped to corresponding transcripts. This approach allows us to maintain consistency of old identifiers and statistics. Linkage of peptides to transcripts was realized by a mapping table, which translates ESTs into all possible open reading frames (ORF). Only experimentally verified peptide sequences that map to an ORF table are included in the database.
Individual sections can be updated separately since, e.g. annotations or ontologies usually change more often than transcript sequences or associated peptide data. The update routine for the annotation section allows different instances as discussed above. Since our functional annotations depend on similarities to a Uniprot protein, features describing a transcript can be extended by any other information appended to the Uniprot identifiers. Our database design allowed us to implement a database frontend with the central object ‘transcript’. Starting from a transcript in Newt-omics, all affiliated data can be directly accessed.
Graphical user interface
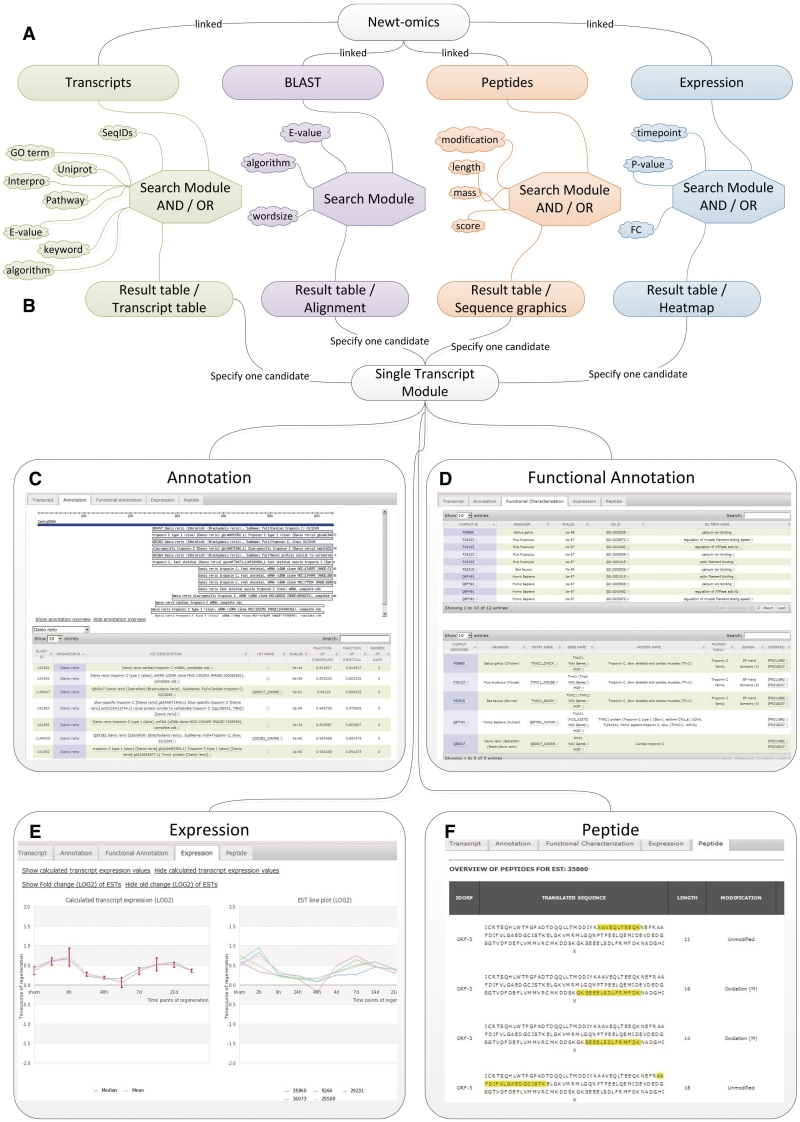
To facilitate easy access to the database we developed a web-based user interface. The user interface was designed using PHP/HTML/JS scripts. Database searches can be initiated from the four main windows ‘Transcripts’, ‘Blast’, ‘Peptides’ and ‘Expression’ (Figure 1A). The main window ‘Transcripts’ contains: (i) a transcript-centred search enabling access to internal EST or transcript identifiers, transcript length and name; (ii) an annotation-centred search allowing selection of the annotation source (algorithm and database) and searches for keywords or significance (e-value); (iii) a functional annotation search allowing searches for GO term numbers, Uniprot Identifiers, pathways, protein families and Interpro domains for single transcripts or a group of transcripts of interest. Complex searches can be performed by a combination of logical AND or OR statements within the three search tabs (Figure 1A). Searches can be filtered by expression changes and by transcripts for which at least one peptide has been experimentally validated. Search results from the ‘Transcripts’ window are displayed in a table, listing transcripts that match to a query (Figure 1B). This results table can be sorted by transcript ID and number, the number of ESTs corresponding to the transcript, the length of the assembled transcript, the number of annotations from a BLAST search and the number of hits in functional annotations. A detailed list, showing all individual annotation results, can be expanded from a tab on the bottom of the results page. Here, the blast alignment can be visualized by moving over the alignment link of an annotation hit. The main window ‘BLAST’ enables similarity searches to any sequence entry in the Newt-omics database using the BLAST tools. Searches in this window can be filtered by e-value, BLAST algorithm and by data source (Figure 1A). Results of a BLAST search are displayed according to NCBI BLAST results, with a graphical coverage, a results list providing direct links to matching transcripts and a sequence alignment view (Figure 1B). The main window ‘Peptides’ permits searches for experimentally validated peptide properties. The form has a query field for core attributes of peptides such as, length, mass, mascot score and modification. These core attributes can be chosen from individual experiments sets or RAW files (Figure 1A). The results table of a peptide search displays matching ESTs, corresponding peptide sequences and peptide properties, including length of the peptide, modification and molecular mass (Figure 1B). The main window ‘Expression’ provides access to microarray expression data from regenerating newt heart. The search form is able to combine queries for fold changes and P-values for each individual time point of the experiment (Figure 1A). Search results are visualized in a heat map, displaying all matching transcripts with their ID and name and expression values with corresponding P-values for each time point of an experiment (Figure 1B).
Figure 1.
The hierarchical structure of the Newt-omics Graphical User Interface (GUI) (A) The four main windows Transcripts BLAST, Peptides, Expression allow searches for single transcripts or a group of transcripts from different affiliated data sources. (B) The results tables list transcripts that link to the ‘Single Transcript Module’. All information linked to a selected transcript can be accessed via the four tabs Annotation (C), Functional Annotation (D), Expression (E) and Peptide (F).
Once a transcript of interest has been identified by any of the four main search windows, its selection directly links to the single ‘Transcript’ module (Figure 1B), which displays the cDNA sequence of a selected transcript and lists the IDs and coordinates of the individual ESTs that comprise the transcript. From this central object, all affiliated information can be accessed via the tabs, ‘Annotation’, ‘Functional Annotation’, ‘Expression’ and ‘Peptide’.
The ‘Annotation’ view lists all information related to similarity searches for the selected transcript. The list can be sorted and selected by source organism, by hit description, corresponding identifiers from public databases, and for statistics of an alignment, like e-value, fraction of conserved, fraction of identical and number of gaps. Further information to a selected transcript is provided by an alignment graphics and an annotation overview that shows all organisms for which a similarity has been identified and displays the proportion of total annotations for each organism (Figure 1C). The ‘Functional Annotation’ view lists and links to GO terms affiliated to an individual transcript. This list can be sorted by organism, e-value, UNIPROT ID, GO identifier and GO term name. A submenu below contains more detailed information about identified protein domains or Interpro crosslinks to UNIPROT identifiers. This list can also be sorted by organisms, by UNIPROT ID and corresponding protein names and abbreviations. It also provides links to wikigenes (24), and iHOP (25), corresponding protein families and protein domains with associated interpro links (Figure 1D).
The ‘Expression’ view provides a detailed view of calculated expression values for a transcript and the individual ESTs, comprising a selected transcript. Graphs of calculated mean and median fold changes with standard deviation are displayed for a transcript on the left, whereas expression changes for individual ESTs comprising the transcript are displayed on the right. Calculated fold changes for the selected transcript and fold changes of the individual ESTs can be displayed in separate tables (Figure 1E).
The ‘Peptide’ view provides detailed information for experimentally validated peptides, corresponding to a selected transcript. Since peptide identifications are based on EST sequences, the matching peptides are highlighted within translated ESTs that constitute a transcript. Detailed information to a peptide including length, modification, modified sequence, Mascot score and mass to charge ratio is listed right to the corresponding peptide (Figure 1F).
DISCUSSION
Newt-omics database is the first integrated repository for the red spotted newt. It constitutes a reference resource for transcripts and proteins that are expressed in the newt heart. Furthermore, it allows a detailed gene expression analysis of newt heart regeneration. The database scheme was developed with a focus on (i) pre-processing of biological data and bioinformatics analysis, (ii) extensibility and (iii) modelling of biological data. Our approach features a user-friendly web interface, which allows intuitive access for researchers with limited bioinformatical training.
For high content data sets that are based on large batch sequencing or microarray time course experiments, appropriate bioinformatics is crucial for the generation of useful annotations and functional assignments. Raw data are usually of limited value for non-bioinformaticians. Therefore, we decided to store only bioinformatically pre-processed data sets in Newt-omics. Similarly, we wanted to visualize the quality of our bioinformatics analysis by graphical representation, rather than giving statistical values, which facilitates access to non-specialists. For example, data plots visualize the uniformity of transcript expression during the time course of heart regeneration. Line plots for individual members of a transcript help to identify irregular EST expression patterns that might arise due to imperfect sequence assembly or by alternative splice isoforms. The graphical peptide view identifies positions of peptides within an ORF, that allows discrimination between coding and non-coding areas of a transcript. This feature is extremely helpful to analyse transcripts that lack any sequence homology, since the existence of matching peptides identify transcripts as true protein coding. Since we performed homology searches on multiple organisms and source databases, it is possible to sort identified sequences by source organism, significance of similarity and description. Such a sorting approach provides information about the degree of conservation between multiple species. Likewise, involvement of molecules in regenerative processes known from other organisms might be uncovered by their functional annotations. Newt-omics allows identification of GO terms of interest that are associated with tissue development or regeneration in mouse, human, fish and chicken.
Another important feature of our database is the ability to update each part of the database separately without disturbing the integrity of the database. It is evident that newly emerging sequencing technologies will make traditional Sanger-based sequencing approaches of new model organisms obsolete even if no ‘matrix’ for assembly sequence reads is available. Although recent algorithms for de novo assemblies of short reads are still challenging, the rapid development of this field will help to overcome existing obstacles. Moreover, the Sanger-sequencing reads stored in our database provide a solid basis for future assembly projects and facilitate gene expression experiments based on arrays and short read sequencing. Future updates of the database will include a more comprehensive set of transcriptome sequencing data derived from other tissues than regenerating heart. This data set will also allow to increase the number of peptides that can be identified from mass spectrometry measurements.
The design of our database emphasizes the linkage between the different data sets. The web-based frontend enables an intuitive ‘from each view to any other view’ within the database. We have focused primarily on the ‘transcript’ as the central object of the database, which might help to address biologically relevant processes. In contrast, the central element of other relational databases with a similar approach relies on genome information (26), which is more difficult to link to biological processes.
Taken together, our database provides molecular insights into a valuable, yet relatively little studied model organism that allows detailed characterization of regenerative processes. The database provides a comprehensive repository not only for researchers working with N. viridescens but also for others in closely related biological disciplines like developmental biology and regenerative medicine. Newt-omics complements large publicly available databases and provides detailed information for research in regenerative medicine ranging from salamanders to humans (27–29).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figure 1.
FUNDING
The Max Planck Society and the Excellence Initiative ‘Cardiopulmonary System’; the University of Giessen-Marburg Lung Centre (UGMLC); the Cell and Gene Therapy Centre (CGT) supported by the HMWK. Funding for open access charge: Max Planck Institute for Heart and Lung Research.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We wish to thank Sven Klages from the Max Planck Institute for Molecular Genetics Berlin for the initial sequence processing and his helpful suggestions concerning annotation of data. We also want to thank Matthew Wheeler from the Max Planck Institute for Heart and Lung Research Bad Nauheim for his helpful suggestions concerning the manuscript.
REFERENCES
- 1.Bowes JB, Snyder KA, Segerdell E, Gibb R, Jarabek C, Noumen E, Pollet N, Vize PD. Xenbase: a Xenopus biology and genomics resource. Nucleic Acids Res. 2008;36:D761–D767. doi: 10.1093/nar/gkm826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bradford Y, Conlin T, Dunn N, Fashena D, Frazer K, Howe DG, Knight J, Mani P, Martin R, Moxon SA, et al. ZFIN: enhancements and updates to the Zebrafish Model Organism Database. Nucleic Acids Res. 2011;39:D822–D829. doi: 10.1093/nar/gkq1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.de la Cruz N, Bromberg S, Pasko D, Shimoyama M, Twigger S, Chen J, Chen CF, Fan C, Foote C, Gopinath GR, et al. The Rat Genome Database (RGD): developments towards a phenome database. Nucleic Acids Res. 2005;33:D485–D491. doi: 10.1093/nar/gki050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Diez-Roux G, Banfi S, Sultan M, Geffers L, Anand S, Rozado D, Magen A, Canidio E, Pagani M, Peluso I, et al. A high-resolution anatomical atlas of the transcriptome in the mouse embryo. PLoS Biol. 2011;9:e1000582. doi: 10.1371/journal.pbio.1000582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Drysdale R. FlyBase: a database for the Drosophila research community. Methods Mol. Biol. 2008;420:45–59. doi: 10.1007/978-1-59745-583-1_3. [DOI] [PubMed] [Google Scholar]
- 6.Brockes JP. Amphibian limb regeneration: rebuilding a complex structure. Science. 1997;276:81–87. doi: 10.1126/science.276.5309.81. [DOI] [PubMed] [Google Scholar]
- 7.Iten LE, Bryant SV. Regeneration from different levels along the tail of the newt, Notophthalmus viridescens. J. Exp. Zool. 1976;196:293–306. doi: 10.1002/jez.1401960304. [DOI] [PubMed] [Google Scholar]
- 8.Tsonis PA. Amphibian limb regeneration. In Vivo. 1991;5:541–550. [PubMed] [Google Scholar]
- 9.Call MK, Grogg MW, Tsonis PA. Eye on regeneration. Anat. Rec. B New Anat. 2005;287:42–48. doi: 10.1002/ar.b.20084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Henry JJ, Tsonis PA. Molecular and cellular aspects of amphibian lens regeneration. Prog. Retin. Eye Res. 2010;29:543–555. doi: 10.1016/j.preteyeres.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Berg DA, Kirkham M, Beljajeva A, Knapp D, Habermann B, Ryge J, Tanaka EM, Simon A. Efficient regeneration by activation of neurogenesis in homeostatically quiescent regions of the adult vertebrate brain. Development. 2010;137:4127–4134. doi: 10.1242/dev.055541. [DOI] [PubMed] [Google Scholar]
- 12.Parish CL, Beljajeva A, Arenas E, Simon A. Midbrain dopaminergic neurogenesis and behavioural recovery in a salamander lesion-induced regeneration model. Development. 2007;134:2881–2887. doi: 10.1242/dev.002329. [DOI] [PubMed] [Google Scholar]
- 13.Bader D, Oberpriller JO. Repair and reorganization of minced cardiac muscle in the adult newt (Notophthalmus viridescens) J. Morphol. 1978;155:349–357. doi: 10.1002/jmor.1051550307. [DOI] [PubMed] [Google Scholar]
- 14.Borchardt T, Braun T. Cardiovascular regeneration in non-mammalian model systems: what are the differences between newts and man? Thromb. Haemost. 2007;98:311–318. [PubMed] [Google Scholar]
- 15.Oberpriller JO, Oberpriller JC. Response of the adult newt ventricle to injury. J. Exp. Zool. 1974;187:249–253. doi: 10.1002/jez.1401870208. [DOI] [PubMed] [Google Scholar]
- 16.Brockes J, Kumar A. Newts. Curr. Biol. 2005;15:R42–44. doi: 10.1016/j.cub.2004.12.049. [DOI] [PubMed] [Google Scholar]
- 17.Borchardt T, Looso M, Bruckskotten M, Weis P, Kruse J, Braun T. Analysis of newly established EST databases reveals similarities between heart regeneration in newt and fish. BMC Genomics. 2010;11:4. doi: 10.1186/1471-2164-11-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Looso M, Borchardt T, Kruger M, Braun T. Advanced identification of proteins in uncharacterized proteomes by pulsed in vivo stable isotope labeling-based mass spectrometry. Mol. Cell Proteomics. 2010;9:1157–1166. doi: 10.1074/mcp.M900426-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barrell D, Dimmer E, Huntley RP, Binns D, O'Donovan C, Apweiler R. The GOA database in 2009–an integrated Gene Ontology Annotation resource. Nucleic Acids Res. 2009;37:D396–D403. doi: 10.1093/nar/gkn803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kruger M, Moser M, Ussar S, Thievessen I, Luber CA, Forner F, Schmidt S, Zanivan S, Fassler R, Mann M. SILAC mouse for quantitative proteomics uncovers kindlin-3 as an essential factor for red blood cell function. Cell. 2008;134:353–364. doi: 10.1016/j.cell.2008.05.033. [DOI] [PubMed] [Google Scholar]
- 21.Schwanhausser B, Gossen M, Dittmar G, Selbach M. Global analysis of cellular protein translation by pulsed SILAC. Proteomics. 2009;9:205–209. doi: 10.1002/pmic.200800275. [DOI] [PubMed] [Google Scholar]
- 22.Elias JE, Gygi SP. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat. Methods. 2007;4:207–214. doi: 10.1038/nmeth1019. [DOI] [PubMed] [Google Scholar]
- 23.Cheng XY, Chen XH, Hua J. The Overview of Entity Relation Extraction Methods. Comm Com Inf Sc. 2011;134:749–754. [Google Scholar]
- 24.Hoffmann R. A wiki for the life sciences where authorship matters. Nat. Genet. 2008;40:1047–1051. doi: 10.1038/ng.f.217. [DOI] [PubMed] [Google Scholar]
- 25.Hoffmann R, Valencia A. A gene network for navigating the literature. Nat. Genet. 2004;36:664–664. doi: 10.1038/ng0704-664. [DOI] [PubMed] [Google Scholar]
- 26.Zhou P, Emmert D, Zhang P. Using Chado to store genome annotation data. Curr. Protoc. Bioinformatics. 2006 doi: 10.1002/0471250953.bi0906s12. Chapter 9, Unit 9.6. [DOI] [PubMed] [Google Scholar]
- 27.Antos CL, Tanaka EM. Vertebrates that regenerate as models for guiding stem cels. Adv. Exp. Med. Biol. 2010;695:184–214. doi: 10.1007/978-1-4419-7037-4_13. [DOI] [PubMed] [Google Scholar]
- 28.Ausoni S, Sartore S. From fish to amphibians to mammals: in search of novel strategies to optimize cardiac regeneration. J. Cell Biol. 2009;184:357–364. doi: 10.1083/jcb.200810094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Menger B, Vogt PM, Kuhbier JW, Reimers K. Applying amphibian limb regeneration to human wound healing: a review. Ann Plast Surg. 2010;65:504–510. doi: 10.1097/SAP.0b013e3181d376f9. [DOI] [PubMed] [Google Scholar]