Abstract
Recent advances in sequencing technology have created unprecedented opportunities for biological research. However, the increasing throughput of these technologies has created many challenges for data management and analysis. As the demand for sophisticated analyses increases, the development time of software and algorithms is outpacing the speed of traditional publication. As technologies continue to be developed, methods change rapidly, making publications less relevant for users. The SEQanswers wiki (SEQwiki) is a wiki database that is actively edited and updated by the members of the SEQanswers community (http://SEQanswers.com/). The wiki provides an extensive catalogue of tools, technologies and tutorials for high-throughput sequencing (HTS), including information about HTS service providers. It has been implemented in MediaWiki with the Semantic MediaWiki and Semantic Forms extensions to collect structured data, providing powerful navigation and reporting features. Within 2 years, the community has created pages for over 500 tools, with approximately 400 literature references and 600 web links. This collaborative effort has made SEQwiki the most comprehensive database of HTS tools anywhere on the web. The wiki includes task-focused mini-reviews of commonly used tools, and a growing collection of more than 100 HTS service providers. SEQwiki is available at: http://wiki.SEQanswers.com/.
INTRODUCTION
Recent developments in sequencing technology have required wet-lab biologists and bioinformaticians to collaborate like never before. These new technologies are driving the rapid development of new software and algorithms for data analysis. Both biologists and bioinformaticians with a pressing need for computational tools now have a multitude of published software to choose from. However, more choice does not necessarily confer increased productivity. In contrast, choice can be confusing and can impair decision making (1). In addition, users without access to closed-source journals are deprived of opportunities to understand the tools published there.
The rapid emergence of tools for HTS analysis is exceeding the capacity of an individual user, or even an individual institution, to monitor all the developments in the field. Funding often limits the ability of institutions to engage in such non-core activities. Traditionally, practices such as journal clubs have been used to educate researchers on current topics. However, in such a setting, the attendees are often confined to a geographical space, usually within an institution or local region. The limited number of topics per-session impedes knowledge transfer in a rapidly developing field. All these issues call for a robust system for rapid sharing of knowledge.
As international collaboration in the sciences increases (2), community-curated databases and websites are gaining in popularity (3). Previously, ‘wikification’ of primary sequence databases such as GenBank has faced stiff resistance (4). GenBank, as an important archive of sequences and annotations, should be assured of content accuracy. However, in our opinion a small group of curators cannot fully encompass the collective expertise of the larger scientific community (5). Indeed, in order to allow for prompt error correction while maintaining content accuracy, Steven Salzberg suggested a layer of wiki be added to existing expert-curated databases (6). For example, a ‘wiki-track’ has been implemented in the UCSC Genome Browser for community annotation of UCSC genes and genomic locations.
The SEQanswers forum: a credible HTS community
SEQanswers.com was founded as a central hub for technical communication about high-throughput sequencing (HTS) technologies. Consisting primarily of a forum, the SEQanswers community enables rapid dissemination of both wet-lab techniques and information regarding computational tools and analyses. The forum allows new tools, techniques and pipelines to be rapidly announced, tested, benchmarked and discussed within an active community. Over 20 000 registered users composed of a diverse mix of bioinformaticians, geneticists and molecular biologists, meet and share their experiences and tools. The forum is a broad, user-driven resource, focused on all aspects of high-throughput genomics. Participation is open to everyone, regardless of scientific background, institution or level of knowledge. Since its establishment in late 2007, the community has already been cited more than 10 times in high profile journals, including Nature, PLoS and Nucleic Acids Research.
As a supplement to traditional forms of scientific communication, SEQanswers offers instantaneous sharing of ideas, findings and reviews between peers at the cutting edge of high-throughput genomics. The site has become an important resource for worldwide collaboration and education in the modern genomics era. SEQanswers allows interaction among bioinformatics analysts and between users and developers.
Traditional software release, in the form of email announcements or on a tool's website, require the users prior interest in the tool. SEQanswers allows any registered member to announce their own tools, even preliminary ones. Software developers thus reach a larger pool of potential users. Developer feedback via traditional channels is usually private. In contrast, SEQanswers users are encouraged to provide feedback in a public forum. Ideas are rapidly developed, practical and theoretical issues are debated. This process leads to improved software development.
The SEQanswers wiki: a structured wiki database for the SEQanswers community
The SEQanswers wiki (SEQwiki) originated from a long running discussion thread in the SEQanswers forum, where an ever growing list of software was being maintained. Although the forum worked well for discussion, it lacked collaborative editing and categorization of resources. There was a high burden on the forum administrators to structure the information being added to the discussion. The wiki was initially started to distribute and organize this activity.
The description of the wiki is the focus of the remainder of this publication.
SEQwiki
Broadly, the content of SEQwiki falls into three categories: (i) software, (ii) service providers and (iii) tutorials. Each of these three sections is described below.
A structured catalogue of bioinformatics tools
SEQwiki is a structured catalogue of bioinformatics tools for HTS analysis that is actively maintained by the community. Pages for each tool include the tool’s data types, formats, capabilities and provenance. Each page is supplemented by links to publications and online resources. At the time of writing, the wiki provides information on more than 500 unique software tools, including approximately 400 references and 600 web links.
Data is contributed in both structured and free text formats using either forms or regular ‘wiki text’, respectively. The form for adding or editing a tool was created in the wiki using the powerful Semantic Forms extension. In combination with Semantic MediaWiki and other extensions, SEQwiki functions as a database, allowing reports and queries to be issued over the data, and for advanced searching and navigation to be designed (for an example, see Figure 1).
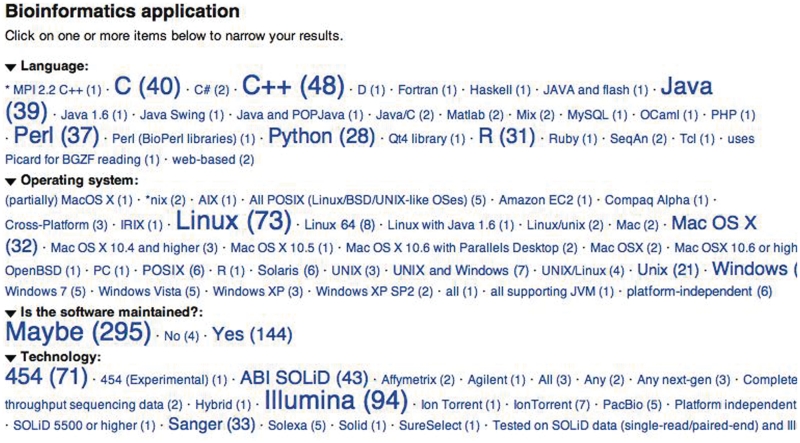
Figure 1.
The ‘Semantic Drilldown’ view of the bioinformatics applications in the wiki. In this screen-shot, values for four of the fields in the database are shown. The size of each value indicates the number of software packages tagged with that value (with the absolute count given in parenthesis). At a glance we can see that: software written in C++ dominates, followed by C, Java and then Perl. Most tools run in Linux, and most software tools are compatible with Illumina and 454 technologies. With just a few clicks, users can extract a list of tools meeting their exact criteria.
A simple form guides the user through the process of adding standardized data for a new bioinformatics application. The form has three main sections as follows:
The summary section collects a short, free-text description of the tool, the main biological application domain(s), the bioinformatics analysis method(s) employed and the type(s) of compatible HTS technologies.
The authorship section collects details about the tool’s authors, their associated institutions and its current status, maintained or not. Status defaults to ‘maybe’ to encourage user review of the available tools.
The technical section collects details about the tool’s licence, file formats, programming language and the required operating system. These are important attributes for tool selection.
Finally, after saving the form, the user can review the page, and add relevant publications and URLs using sub-forms designed for this purpose.
Forms created in the wiki use jQuery based auto-completion to help users to discover existing tools and find common values for the form fields. This helps to enforce data consistency, but doesn’t constrain users to a fixed set of options.
Each tool is ‘tagged’ by the values entered in the various fields. A tool can have multiple values in each field, for example, a program for analysis of RNA sequencing could function as both a read aligner and as a tool to identify splice-junctions. The full list of tools can be narrowed down using multiple parameters. A tool can be discovered in this way as long as any one of its attributes is matched (Figure 1).
The tool pages present the entered data in a table on the right, with the ‘free text’ about the tool appearing on the left. The added links and literature references follow the free text. At the foot of each page, we include a standard table of links that allow the user to search for the tool in a variety of resources, including the SEQanswers forum. By linking back to the forum in this way, users are guided back to community discussion about the tool.
Through tagging, bioinformaticians focusing on real data can quickly retrieve a collection of up-to-date tools for analysis, while tool authors will be able to find the most appropriate tools to benchmark their programs. The database helps to organize the underlying data, and lets users and developers concentrate on data analysis and development rather than finding the right tools for the right job.
The quality and amount of information for each tool depends on the level of community participation. To help promote the improvement of pages for tools with little information, we have implemented a simple ‘article score’. The score increases for every field that is filled in for the description of the tool. Tools with low article scores are prominently displayed on the software hub page, along with the overall size of the article in bytes.
A directory of service providers
SEQwiki hosts detailed information for more than one hundred HTS service providers from around the globe. Service providers are added in the same way as tools, using a specific form. On a summary page, the table of providers is dynamically generated, and is presented using the Exhibit results format. The resulting table of providers can be narrowed using Facets that cover the types of service provided and the geographic region of the provider.
Finding a service provider geographically close to the user is of great importance to HTS users to ensure quick sample transfer and to maintain sample integrity. This section is invaluable for researchers without HTS core facilities in their own institution. Additionally, this section is for those curious about the current deployment of HTS facilities in different geographical locations.
An educational resource
SEQwiki hosts several short, task-oriented articles or tutorials that compare the major tools in the areas of de novo genome assembly, de novo transcriptome assembly, sequence alignment and SNP calling methodologies. These task-oriented pages explain commonly used software along with their advantages, potential problems and technical requirements. The aim of these pages is to provide the community with continuous updates of bioinformatics software tools.
USAGE STATISTICS
SEQwiki has grown steadily throughout its 2-year life (Figure 2). From Figure 2, it is clear that much content has been created during sporadic bursts of activity, but that there has been a trend of continuous growth at a lower level. Most of the largest bursts of activity occurred within the first year, after which growth is slower but still accounts for a significant number of new pages and revisions.
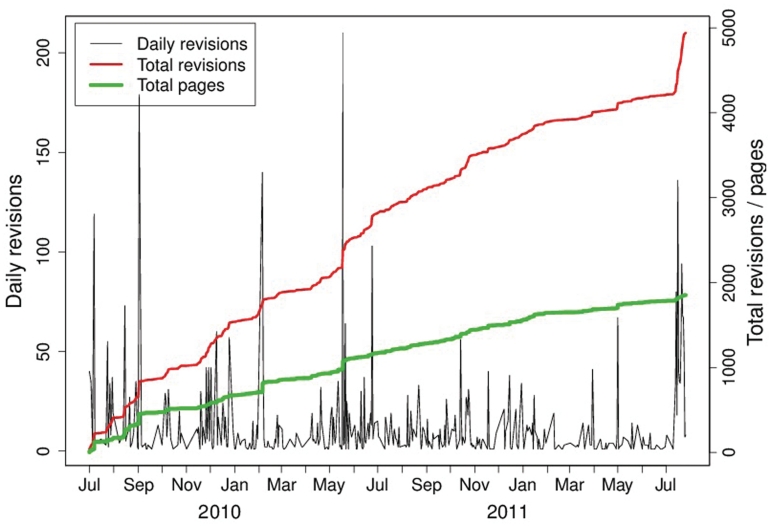
Figure 2.
The growth of SEQwiki since its inception in July 2009 until August 2011. The number of revisions per day (black) varies wildly during the 2-year period, from zero on many days to more than 100 on some days. The cumulative number of revisions is shown as the upper (red) trend. The lower green trend shows the cumulative number of pages in the wiki. Note that one ‘revision’ could be either an edit to an existing page or the creation of a new page.
The pattern of growth in Figure 2 is consistent with the activity of a few ‘super-users’ who make many edits or add many pages against a background of a much larger number of contributors who typically make very few edits (7). This observation is clearly shown again in Figure 3.
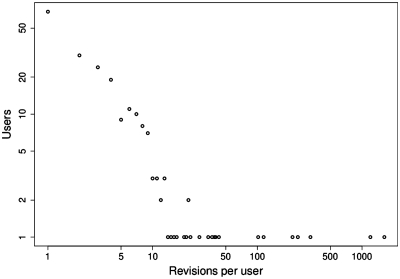
Figure 3.
The heavy tail of SEQwiki contributors. Most contributors make just one or only a few revisions, i.e. 68 users have only ever made one revision. In contrast, a few ‘super-users’ have made hundreds of revisions.
FUTURE DIRECTIONS
SEQwiki is an on-going project. The number of records in the wiki will continue to increase as new software tools are announced and published. The wiki has served as a successful platform providing searching capabilities on software tools for users. We plan to further enhance the features of the wiki. In particular, we would like to improve the following areas:
Community review system: before SEQanswers, user feedback was almost exclusively directed to software authors. Only a few disparate, well established or well-funded groups provided publicly archived mailing lists for community discussion. Software reviews by bloggers were similarly posted independently and relatively rarely. SEQanswers has fostered lively review of both pre-publication and post-publication tools. Usually, long before peer review publication, tools have been announced in SEQanswers and tested extensively within the community. Post-publication improvement and benchmarking among developers is encouraged by discussions in the SEQanswers forum. SEQwiki currently provides a quality metric by presenting the total view count of a tool’s page and the citation count of its associated publications. In future, functionality to provide detailed ratings and reviews could be added to the wiki.
Community-wide benchmarking: Systematic benchmarking is a very important task that is rarely achieved. A few community-wide HTS benchmarking projects are currently underway, including the Critical Assessment of Genome Interpretation (CAGI), the Assemblathon (8), Genome Assembly Gold-Standard Evaluations (CAGE) and dbGASP. By using the wiki, we aim to distribute and ‘crowd source’ this type of data directly from the community. Data sets for benchmarking software tools could be hosted, and performance results could be collected by the community. Software developers would be motivated to provide information on their tools, and users would be motivated to feed back the results of their important individual and hitherto largely unpublishable experimentation.
Integration with the semantic web: there are several efforts to organize databases and software in the life sciences using ontologies, for example, the Software Ontology and the EDAM Ontology. The keywords describing software tools in SEQwiki could be organized using these controlled vocabularies. These data would be distributed over the semantic web using standards such as Resource Description Framework (RDF), which is integrated into Semantic MediaWiki. This approach allows reuse of data either directly on external web pages or by external tools that wish to query SEQwiki. Similarly, data could be queried from the semantic web and presented within SEQwiki.
CONCLUSION
It has long been thought that wikis can play an important role for community annotation in the life sciences (9). An exhaustive list of BioWikis is available at The Bioinformatics Organization.
SEQwiki is a community maintained wiki database that serves as a rapid, day-to-day reference for practitioners in the HTS field. The database refers users to tools and their publications. Community knowledge of tools is extensively discussed in the SEQanswers forum.
SEQwiki is open to edit by an active community around the world. Registered members can submit and curate profiles of bioinformatics software tools. Similar to Wikipedia each modification in the wiki is associated with a registered user, and can be reverted if faulty information is found. Thanks to the active participation of the community, the wiki has met our goals to collect and categorize the ever growing list of bioinformatics software for HTS analysis.
ELECTRONIC ADDRESSES
The discussion where SEQwiki originated: http://seqanswers.com/forums/showthread.php?t=43
The Semantic MediaWiki extension: http://semantic-mediawiki.org/wiki/Semantic_MediaWiki
The Semantic Forms extension: http://www.mediawiki.org/wiki/Extension:Semantic_Forms
The Exhibit results format: http://www.mediawiki.org/wiki/Extension:Semantic_Result_Formats
Software Ontology: http://www.ebi.ac.uk/efo/swo
EDAM Ontology: http://edamontology.sourceforge.net/
BioWikis page at The Bioinformatics Organization: http://www.bioinformatics.org/wiki/BioWiki
Critical Assessment of Genome Interpretation: http://genomeinterpretation.org/
Genome Assembly Gold-Standard Evaluations: http://gage.cbcb.umd.edu/
dbGASP: http://cnag.bsc.es/
FUNDING
SEQanswers Community. Funding for open access charge: Waived by Oxford University Press.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors would like to thank everyone in the SEQanswers community, and everyone who contributed to the creation and curation of the wiki. DMB would like to thank ECO and KR for initial support of SEQwiki and JWL for writing the first draft of the manuscript.
REFERENCES
- 1.Iyengar SSL, Mark R. When choice is demotivating: can one desire too much of a good thing? J. Pers. Soc. Psychol. 2000;79:995–1006. doi: 10.1037//0022-3514.79.6.995. [DOI] [PubMed] [Google Scholar]
- 2.Society TR. Knowledge, Networks and Nations: Global Scientific Collaboration in the 21st Century. Carlton House Terrace, London: The Royal Society; 2011. [Google Scholar]
- 3.Butler D. Science in the web age: joint efforts. Nature. 2005;438:548–549. doi: 10.1038/438548a. [DOI] [PubMed] [Google Scholar]
- 4.Pennisi E. Proposal to ‘Wikify’ GenBank meets stiff resistance. Science. 2008;319:1598–1599. doi: 10.1126/science.319.5870.1598. [DOI] [PubMed] [Google Scholar]
- 5.Hu JC, Aramayo R, Bolser D, Conway T, Elsik CG, Gribskov M, Kelder T, Kihara D, Knight TF, Pico AR, et al. The emerging world of wikis. Science. 2008;320:1289–1290. doi: 10.1126/science.320.5881.1289b. [DOI] [PubMed] [Google Scholar]
- 6.Salzberg S. Genome re-annotation: a wiki solution? Genome Biol. 2007;8:102. doi: 10.1186/gb-2007-8-1-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kittur A, Chi EH, Pendleton BA, Suh B, Mytkowicz T. Power of the few vs. wisdom of the crowd: Wikipedia and the rise of the bourgeoisie. In alt.CHI at 25th Annual ACM Conference on Human Factors in Computing Systems (CHI 2007) 2007 2007 April 28 – May 3. San Jose, CA. [Google Scholar]
- 8.Earl DA, Bradnam K, St John J, Darling A, Lin D, Faas J, Yu HO, Vince B, Zerbino DR, Diekhans M, et al. Assemblathon 1: a competitive assessment of de novo short read assembly methods. Genome Res. 2011 doi: 10.1101/gr.126599.111. September 16 (doi:10.1101/gr.126599.111; epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Waldrop M. Big data: Wikiomics. Nature. 2008;455:22–25. doi: 10.1038/455022a. [DOI] [PubMed] [Google Scholar]