Abstract
The importance of binding site plasticity in protein–ligand interactions is well-recognized, and so are the difficulties in predicting the nature and the degree of this plasticity by computational means. To assist in understanding the flexible protein–ligand interactions, we constructed the Pocketome, an encyclopedia of about one thousand experimentally solved conformational ensembles of druggable binding sites in proteins, grouped by location and consistent chain/cofactor composition. The multiplicity of pockets within the ensembles adds an extra, fourth dimension to the Pocketome entry data. Within each ensemble, the pockets were carefully classified by the degree of their pairwise similarity and compatibility with different ligands. The core of the Pocketome is derived regularly and automatically from the current releases of the Protein Data Bank and the Uniprot Knowledgebase; this core is complemented by entries built from manually provided seed ligand locations. The Pocketome website (www.pocketome.org) allows searching for the sites of interest, analysis of conformational clusters, important residues, binding compatibility matrices and interactive visualization of the ensembles using the ActiveICM web browser plugin. The Pocketome collection can be used to build multi-conformational docking and 3D activity models as well as to design cross-docking and virtual ligand screening benchmarks.
INTRODUCTION
The biological machinery relies on transient intermolecular interactions as the main communication tool. The sites of protein interactions with endogenous small molecules and peptides are of particular interest because they are also often binding sites for therapeutic or toxic chemicals and their metabolites. The inherent flexibility of such binding sites is of primary biological importance because it allows them to accommodate a variety of binding partners; however, it also often makes it difficult or even impossible to predict or rationalize some of the interactions (1–7).
Here, we present the Pocketome, a comprehensive yet clean collection of conformational ensembles of all druggable binding sites, which can be identified experimentally from co-crystal structures in the Protein Data Bank [PDB (8)]. The Pocketome philosophy, first presented in (1), is based on the understanding that some sites on the surface of biopolymers or their permanent assemblies possess the ability to specifically and efficiently form transient complexes with diverse molecular partners, accommodating them through conformational changes of varying degree. A 3D structure of a single complex gives only a limited static view of this functionality; however, thorough cataloging, classification and annotation of the multiple snapshots at each individual site adds the fourth dimension to the data (9), which not only allows separation of spurious or permanent complexes from truly relevant transient interactions, but also provides valuable insights into mechanisms and principles of these interactions.
The focus on the concept of a conformationally variable binding site is the main feature that distinguishes the Pocketome from other existing online databases that collect, enrich and make inferences from the PDB structures of protein complexes with small chemicals: PCIDB (10), MOAD (11), IBIS (12) or ReliBase (13). The Pocketome approach shares some similarity with those of PCDB (14), PepX (15) or DIMA (16), though with specific focus on structural details of the interaction sites. The Pocketome employs a unique algorithm that, in addition to simple binary protein–ligand interactions, enables automatic identification of sites located at multimer assembly interfaces or containing bound cofactors and metal ions, and efficiently separates the sites into permanent and variable parts. The subsequent processing of the Pocketome ensembles creates accurate ligand–residue interaction maps, quantifies cross-compatibility between pockets and ligands from different structures, and performs their conformational clustering.
The Pocketome encyclopedia can assist elucidation of the conserved determinants of molecular interactions, understanding the effects of SNPs and single-point mutations, explanation of protein flexibility and induced fit phenomena, and development of flexible docking algorithms. Moreover, it may become the foundation of structure-based prediction of novel activities of existing molecules, or a tool for activity and binding mode prediction of the new chemical compounds (17–19). With its unique interface providing intuitive but versatile interactive molecular visualization, the Pocketome is a valuable resource for biological, chemical and computational communities in understanding biological function and molecular interactions directly from the structural perspective.
POCKETOME CONTENT
Concepts and terminology
In the Pocketome encyclopedia, the following hierarchy of concepts is assumed. A ‘protein’ is an entity described by a unique invariable sequence and corresponding to an entry in the reviewed part of the Uniprot Knowledgebase (20). Each protein contains one of more structural ‘domains’. A domain has one or more binding ‘sites’, defined as groups of residues binding small ligands. Potential multiplicity of binding sites not only for a single protein, but also for each domain within a single protein is a concept rarely acknowledged (21) but critical for Pocketome logic.
Each Pocketome entry describes a collective binding site and consists of an ensemble of experimentally determined structures of that site, apo or in complex with ligands, superimposed in space, with associated textual and graphical information. The site includes one or several protein chains, and may comprise cofactors and metals. Every ensemble member structure has individual characteristics such as the set of pocket atoms and its own bound ligand.
In Pocketome, we introduced a distinction between the terms ‘pocket’ and ‘site’, with ‘pocket’ describing the sets of atoms that are in direct contact with the cognate ligand in a single experimentally determined complex structure, and ‘site’ being a superset of all pockets projected onto the amino acid sequence. The term ‘site’ therefore describes the set of residues that have been experimentally shown to participate in ligand binding in at least one of the complexes.
The Pocketome data hierarchy can be illustrated by the following example: the human Abl kinase is a protein represented by ABL1_HUMAN entry in the Uniprot KB. It contains a SH2 domain, a SH3 domain, and a protein kinase domain. On the protein kinase domain, there are two crystallographically characterized small-molecule binding sites, called the ATP site and the myristoyl site (22,23). Each of these sites is represented by a single Pocketome entry consisting of a superimposed structure ensemble, ligand ensemble and associated textual and graphical information. Structures within the ATP-site entry differ by the nature of the bound ligand and the precise pocket, i.e. specific atoms making contacts with the ligand in each case. The set of all ligands found in the ATP-site of Abl crystals make the ligand ensemble for this entry.
Pocketome entry organization
Binding sites initially originate from a set of residues in a single protein domain; however, it is often incorrect to assume that they consist of these residues only. The common situations include:
sites located at subunit interface in a multimeric assembly;
sites partially formed by a cofactor molecule such as NAD or ATP; and
sites that contain one or more coordinated metal ions.
In the Pocketome, these situations are properly analyzed and recorded with several important assumptions that are described in the following paragraphs.
In multimeric assembly interface sites, at most two different proteins are allowed. For simplicity, they are referenced as A (for the main entry protein) and B (for potential hetero-multimetric partner). This requirement does not limit the number of polypeptide chains that form the site, because the chains may be multiple copies of the same protein. Possible architectures of the binding site include homodimers (AA), heterodimers (AB), homotrimers (AAA) and higher order assemblies (AAAA, AAB, ABB, AABB, AAABBB, etc.). Pocketome site architecture may or may not coincide with the structure of the protein biological unit. For example, influenza virus neuraminidase is known to function as a homo-tetramer; however, its tetrameric structure can potentially bind to four different molecules of the ligand (sialic acid or a competitive inhibitor), with each binding site formed by only one of the subunits. In contrast, a single binding site in the homotetrameric HMG CoA reductase is formed by two units of the tetramer. In these two examples, the multimeric architecture of a Pocketome entry represents a subset of that of the true biological assembly. In principle, the opposite situation is also possible, when the Pocketome architecture exceeds the biological assembly. For example, despite the monomeric nature of a protein, the binding for multiple small molecules may be formed at the interface with its crystallographic partner, which ‘dimerizes’ with the main protein molecule in an orientation consistent through the entire ensemble. If such crystallographic partner provides a significant fraction of ligand interactions, we consider it as a part of the binding site. PDB BioMT records are not used in generation of the Pocketome entries.
For cofactor-containing sites, the cofactor is defined as a molecule that binds concurrently (non-competitively) with the ligands and provides some fraction of the ligand interactions in multiple binding site structures. In most cases, Pocketome ‘cofactors’ are also classified as cofactors by other sources such as Uniprot or BRENDA (24) databases. However, the opposite is not true, because in case of competitive binding (e.g. ATP-competitive ligands in protein kinases), the cofactor is not classified as such in the Pocketome. Alternatively, a part of the binding pocket can be consistently formed by a relatively short peptide (e.g. the GRPRTTSFAE substrate peptide in Akt kinase structures), in which case such peptide is also classified as a ‘cofactor’. It is useful to think about a Pocketome ‘cofactor’ as a molecule that must be present in the binding site when cross-docking ligands to pockets between the different site structures.
Metal ions or higher order metal containing clusters (e.g. iron–sulfur clusters) are classified as a part of the Pocketome-binding site if they are consistently present across the multiple structures of that site.
Water molecules are deleted from the current versions of the Pocketome entries.
Within each Pocketome entry, a ‘ligand ensemble’ is found which in general case can contain drug-like small molecules, ATP or other entities that bind competitively with these molecules, peptides of different lengths, proteins or nucleic acids. Small molecule ligands, short peptides (up to 12 amino acids in length) or short nucleotide sequences are included in the ligand ensemble entirely, while for longer polypeptide or nucleotide chains, only the part in direct contact with the pocket is counted as a ligand. In some of the pockets within the ensemble, two or more molecules may concurrently occupy the space that contains a single ligand molecule in other pockets; these molecules are classified as a single composite ligand. By analogy with the Pocketome ‘cofactor’, it is convenient to think about a Pocketome ‘ligand’ as one or more molecules that should be removed from the site when cross-docking a ligand from a different site structure.
Summarizing the above considerations, a generic Pocketome site has the following components:
protein chains of no more than two types (A1, A2, … , B1, B2, … etc.);
cofactors; and
structural or catalytic metal ions.
The multiple pocket structures inside the Pocketome ensemble are complexes of this invariable site with various ligands in the ligand ensemble. They capture the conformational variations induced by either binding of the ligands or changing conditions of the experiment. The files in the online version of the database are simplified to include only non-redundant set of pocket compositions (i.e. distinct ligands in combinations with distinct point mutants only). The full Pocketome entry files containing all related structures are available on request.
Pocketome data flow and filtering criteria
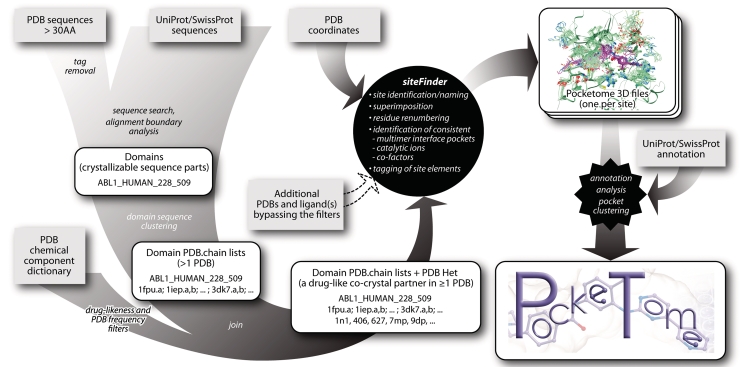
The heart of the Pocketome is the so-called ‘siteFinder’ algorithm that automatically collects, clusters, analyzes and validates the binding pocket structures based on consistency of their composition and spatial configuration between the multiple members of the structural ensemble. A series of additional utilities collect the input for siteFinder by scanning the current releases of the main source databases, PDB and Uniprot (Figure 1). In this process, highly homologous (≥94% sequence identity) proteins originating from different organisms can sometimes be merged into a single entry. This helps to avoid the unnecessary fragmentation of the set and to increase the structural content of the individual ensembles.
Figure 1.
Pocketome data flow. The central part of the pipeline is the siteFinder algorithm. The input for it is generated automatically (by joining several major external databases) or manually. The output of the siteFinder utility in the form of tagged 3D ensembles forms the core of the Pocketome encyclopedia.
As a result of the input generation procedure, each Pocketome entry corresponds to a small-molecule binding site in a protein that (i) has an entry in the reviewed part of the UniProt Knowledgebase, (ii) has been co-crystallized in complex with at least one drug-like small molecule and (iii) is represented in at least two PDB entries. Binding sites that do not satisfy all three of the above requirements may not be represented in the Pocketome collection. This includes:
singletons: sites with only one structure in the PDB;
sites with no co-crystallized drug-like molecules;
variable content/combinatorial sites (e.g. MHC Classes 1 and 2, antibodies, etc.); and
sites in proteins within the unreviewed part of Uniprot.
For selected interesting binding sites that do not pass the filters of the input generation utility, the Pocketome entries can be obtained by providing the siteFinder input manually. For example, this allows construction of the Pocketome entries in cases when there are no structures with drug-like ligands, or when the protein is only described in the unreviewed part of Uniprot.
Because both PDB and Uniprot databases are constantly expanding and developing, and because the Pocketome is updated automatically with their releases, currently excluded entries may become part of the database in future versions.
Pocketome statistics
The current release of the Pocketome encyclopedia (August 2011) contains 988 entries, of which more than 450 (47%) originate from humans and other mammals. Clinically validated drug target classes are represented in the set proportionally to their structural coverage in PDB; for example, the set contains 93 protein kinases, 30 nuclear receptors and 6 G-protein coupled receptors. Each full Pocketome entry contain between 2 and 160 pocket structures (median 14); the non-redundant online entry versions contain between 1 and 40 structures (median 5).
POCKETOME ACCESS
Search options
The online version of the Pocketome encyclopedia is available at http://www.pocketome.org. In the current release, this resource allows search by:
protein Uniprot ID;
protein names, synonym or family;
PDB ID of the structure; and
PDB Hetero ID of the ligand molecule
The searches by protein sequence in FASTA format, ligand SMILES string or drug name/synonym are currently under development and will be added in the future releases of the Pocketome.
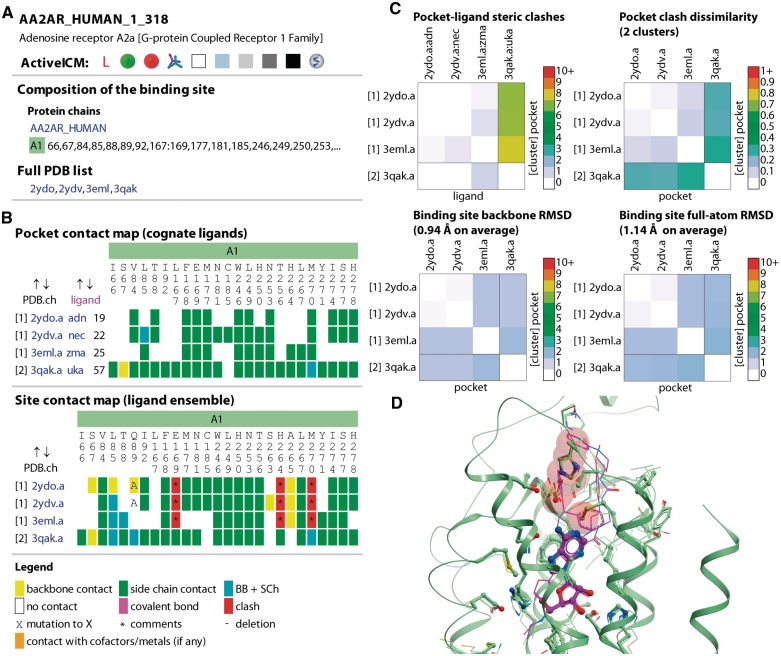
The search results are always returned in the form of a hit list where each hit is represented by the site identifier, protein name and family, the list of all associated PDB entries and the list of PDB Hetero IDs for the ligands in the site. The hits are sorted in the order of decreasing relevance to the search query. In cases when no Pocketome entry exists, some distantly relevant hits may be returned. Clicking on the site identifier takes the user to the individual entry page with ligand–protein contact analysis, pocket pairwise comparison, and interactive visualization of the site ensemble (Figure 2).
Figure 2.
Summary of a single Pocketome page content exemplified by the entry for adenosine A2A receptor. The snapshots were made in August 2011, when four structures of this protein with various modulators were available in PDB (29–31). (A) Summary information: binding site composition (a single polypeptide chain, A1, representing the Uniprot entry AA2AR_HUMAN), list of binding site residues, full PDB list with links. (B) Pocket contact map allows quick identification of conserved (e.g. F168) or ligand-specific (e.g. C185 for NECA, PDB 2ydv) residue contacts; site contact map highlights in red the residues that require significant rearrangement to accommodate at least one of the ligands. (C) Pairwise pocket comparison: the conformation from a co-crystal with agonist UK-432097 (PDB 3eml) is significantly different from the other three, giving rise to two sterically distinct clusters. (D) A snapshot of the ActiveICM ensemble visualization window: NECA-bound conformation appears sterically incompatible with UK-432097 because of multiple rearrangements in the extracellular part of the protein; clashing residues highlighted in the site contact map in panel (B) are shown as transparent red spheres.
Ligand–pocket contact analysis
Two of the three tabs in the text frame of a Pocketome entry are named ‘Pocket’ and ‘Site’, respectively. In addition to the summary information on the pocket/site composition, both tabs present the results of analysis of ligand contacts with the neighboring protein residues and other molecules in the site (Figure 2B). We consider two atoms being in contact if their centers are located at the distance that does not exceed the sum of their van der Waals radii by >20%. A residue is said to be in contact with the ligand if at least one non-hydrogen atom of the ligand contacts this residue. In the ‘Pocket’ tab, the contacts of each individual ligand with its cognate binding pocket are described, allowing quick identification of conserved or ligand-specific interactions. In contrast, the ‘Site’ tab summarizes the ligand ensemble information, most importantly, the contacts that other ligands from the ensemble could make if they were placed into each selected pocket structure. Contacts are presented in compact matrix form and conveniently color coded as backbone only, side-chain only or both. Because ligands from other structures may be sterically incompatible with a selected pocket, the ‘Site’ tab uses an additional type of contact not present in the ‘Pocket’ tab, a ligand–residue ‘steric clash’. A clashing residue is colored red with the color intensity corresponding to the number of ligands spatially overlapping with that residue.
Cases of residue mutations from the reference Uniprot sequence, as well as cases of residue covalent modifications, deletions, or insertions are marked in the ‘Pocket’ and ‘Site’ contact matrices. The matrices are interactively clickable leading to changes in the 3D graphics window on the right.
Pairwise comparison of binding pockets
Understanding binding site flexibility and induced fit effects is one of the primary goals of the Pocketome encyclopedia. Therefore, each Pocketome entry is complemented by a summary page that presents the results of pairwise comparison of the ensemble members by several independent criteria (Figure 2C, ‘Pairwise Comparison’ tab in the entry pages). The two most straightforward plots show the pairwise backbone and full-atom RMSD (root mean square deviation) between the pockets. This RMSD gives an idea about the degree of the conformational variability of the site, however, it is generally considered to be a suboptimal measure for protein structure comparison (25). Therefore, we also compared pockets in terms of their steric compatibility with various ligands in the ensemble. For that comparison, (i) each pocket was described by a vector of steric clashes that it makes with the ligand ensemble, (ii) pocket distances were calculated as normalized absolute difference between the vectors and (iii) all pockets within the entry were clustered according to the calculated distance to form sterically cross-compatible subsets. A total of 300 entries in the Pocketome contain more than one steric cross-compatibility cluster, with the most notable examples including agonist- and antagonist-specific conformations of nuclear receptors (e.g. estrogen receptor α) and active and inactive conformations of protein kinase domains (e.g. Abl).
ActiveICM visualization
Pocketome employs the ActiveICM (26) technology for the interactive visualization of the binding site ensembles (Figure 2D). ActiveICM is a freely available plug-in for all major web browsers with unique capabilities not accessible in other similar tools. ActiveICM version 1.1-6 or higher is required to access all the functionality of the Pocketome pages. ActiveICM is widely accepted and used by the computational and structural biology communities as an advanced format for 3D data visualization (27,28). In the Pocketome pages, the molecular entities forming the binding site are consistently color coded according to their type (e.g. polypeptide chains A1, A2, … , B1, B2, … , cofactors, metal ions, or the ligands) and are presented in several convenient and informative views which are controlled by the links on the left side of the screen, in the ‘Pocket’ and ‘Site’ tabs. Generic ActiveICM controls enable view manipulations like rotation, zooming in and out, centering on selected parts, clipping, rocking, etc. An added advantage of using the ActiveICM technology and data format is that the Pocketome pages are viewable on all Apple portable devices using the sister iMolview application.
FUNDING
This work was partially supported by National Institutes of Health (grant numbers R01 GM071872, U01 GM094612, U54 GM094618 and RC2 LM010994). Funding for open access charge: National Institutes of Health (grant number R01 GM071872).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
Authors thank Max Totrov and Eugene Raush for their help with data processing and visualization, Manuel Rueda for valuable suggestions, Chris Edwards for web server administration and maintenance, and Karie Wright and Marco Neves for critically reading the manuscript.
REFERENCES
- 1.Abagyan R, Kufareva I. The flexible pocketome engine for structural chemogenomics. In: Jacoby E, editor. Chemogenomics: Methods and Applications. Vol. 575. Humana Press; 2009. pp. 249–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nabuurs SB, Wagener M, de Vlieg J. A flexible approach to induced fit docking. J. Med. Chem. 2007;50:6507–6518. doi: 10.1021/jm070593p. [DOI] [PubMed] [Google Scholar]
- 3.de Graaf C, Rognan D. Selective structure-based virtual screening for full and partial agonists of the β2 adrenergic receptor. J. Med. Chem. 2008;51:4978–4985. doi: 10.1021/jm800710x. [DOI] [PubMed] [Google Scholar]
- 4.de Graaf C, Rognan D. Customizing G Protein-coupled receptor models for structure-based virtual screening. Curr. Pharm. Des. 2009;15:4026–4048. doi: 10.2174/138161209789824786. [DOI] [PubMed] [Google Scholar]
- 5.Irwin JJ, Shoichet BK, Mysinger MM, Huang N, Colizzi F, Wassam P, Cao Y. Automated Docking Screens: A Feasibility Study. J. Med. Chem. 2009;52:5712–5720. doi: 10.1021/jm9006966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Katritch V, Reynolds KA, Cherezov V, Hanson MA, Roth CB, Yeager M, Abagyan R. Analysis of full and partial agonists binding to beta2-adrenergic receptor suggests a role of transmembrane helix V in agonist-specific conformational changes. J. Mol. Recognit. 2009;22:307–318. doi: 10.1002/jmr.949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kufareva I, Abagyan R. Type-II kinase inhibitor docking, screening, and profiling using modified structures of active kinase states. J. Med. Chem. 2008;51:7921–7932. doi: 10.1021/jm8010299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rose PW, Beran B, Bi C, Bluhm WF, Dimitropoulos D, Goodsell DS, Prlic A, Quesada M, Quinn GB, Westbrook JD, et al. The RCSB Protein Data Bank: redesigned web site and web services. Nucleic Acids Res. 2011;39:D392–D401. doi: 10.1093/nar/gkq1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bottegoni G, Kufareva I, Totrov M, Abagyan R. Four-dimensional docking: a fast and accurate account of discrete receptor flexibility in ligand docking. J. Med. Chem. 2009;52:397–406. doi: 10.1021/jm8009958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kalinina OV, Wichmann O, Apic G, Russell RB. Combinations of Protein-Chemical Complex Structures Reveal New Targets for Established Drugs. PLoS Comput. Biol. 2011;7:e1002043. doi: 10.1371/journal.pcbi.1002043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Benson ML, Smith RD, Khazanov NA, Dimcheff B, Beaver J, Dresslar P, Nerothin J, Carlson HA. Binding MOAD, a high-quality protein-ligand database. Nucleic Acids Res. 2008;36:D674–D678. doi: 10.1093/nar/gkm911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shoemaker BA, Zhang D, Thangudu RR, Tyagi M, Fong JH, Marchler-Bauer A, Bryant SH, Madej T, Panchenko AR. Inferred Biomolecular Interaction Server: a web server to analyze and predict protein interacting partners and binding sites. Nucleic Acids Res. 2010;38:D518–D524. doi: 10.1093/nar/gkp842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Günther J, Bergner A, Hendlich M, Klebe G. Utilising structural knowledge in drug design strategies: applications using relibase. J. Mol. Biol. 2003;326:621–636. doi: 10.1016/s0022-2836(02)01409-2. [DOI] [PubMed] [Google Scholar]
- 14.Juritz EI, Alberti SF, Parisi GD. PCDB: a database of protein conformational diversity. Nucleic Acids Res. 2011;39:D475–D479. doi: 10.1093/nar/gkq1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vanhee P, Reumers J, Stricher F, Baeten L, Serrano L, Schymkowitz J, Rousseau F. PepX: a structural database of non-redundant protein-peptide complexes. Nucleic Acids Res. 2010;38:D545–D551. doi: 10.1093/nar/gkp893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Luo Q, Pagel P, Vilne B, Frishman D. DIMA 3.0: Domain Interaction Map. Nucleic Acids Res. 2011;39:D724–D729. doi: 10.1093/nar/gkq1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bottegoni G, Rocchia W, Rueda M, Abagyan R, Cavalli A. Systematic Exploitation of Multiple Receptor Conformations for Virtual Ligand Screening. PLoS ONE. 2011;6:e18845. doi: 10.1371/journal.pone.0018845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carlsson J, Yoo L, Gao Z-G, Irwin JJ, Shoichet BK, Jacobson KA. Structure-Based Discovery of A2A Adenosine Receptor Ligands. J. Med. Chem. 2010;53:3748–3755. doi: 10.1021/jm100240h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Katritch V, Jaakola V-P, Lane JR, Lin J, Ijzerman AP, Yeager M, Kufareva I, Stevens RC, Abagyan R. Structure-Based Discovery of Novel Chemotypes for Adenosine A2A Receptor Antagonists. J. Med. Chem. 2010;53:1799–1809. doi: 10.1021/jm901647p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.The UniProt C. Ongoing and future developments at the Universal Protein Resource. Nucleic Acids Res. 2011;39:D214–D219. doi: 10.1093/nar/gkq1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meslamani J, Rognan D, Kellenberger E. sc-PDB: a database for identifying variations and multiplicity of ‘druggable’ binding sites in proteins. Bioinformatics. 2011;27:1324–1326. doi: 10.1093/bioinformatics/btr120. [DOI] [PubMed] [Google Scholar]
- 22.Fabbro D, Manley PW, Jahnke W, Liebetanz J, Szyttenholm A, Fendrich G, Strauss A, Zhang J, Gray NS, Adrian F, et al. Inhibitors of the Abl kinase directed at either the ATP- or myristate-binding site. Biochim. Biophys. Acta Prot. Proteomics. 2010;1804:454–462. doi: 10.1016/j.bbapap.2009.12.009. [DOI] [PubMed] [Google Scholar]
- 23.Iacob RE, Pene-Dumitrescu T, Zhang J, Gray NS, Smithgall TE, Engen JR. Conformational disturbance in Abl kinase upon mutation and deregulation. Proc. Natl Acad. Sci. USA. 2009;106:1386–1391. doi: 10.1073/pnas.0811912106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Scheer M, Grote A, Chang A, Schomburg I, Munaretto C, Rother M, Sohngen C, Stelzer M, Thiele J, Schomburg D. BRENDA, the enzyme information system in 2011. Nucleic Acids Res. 2011;39:D670–D676. doi: 10.1093/nar/gkq1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kufareva I, Abagyan R. Methods of protein structure comparison. Homology Modeling: Methods and Protocols. 2012 doi: 10.1007/978-1-61779-588-6_10. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Raush E, Totrov M, Marsden BD, Abagyan R. A New Method for Publishing Three-Dimensional Content. PLoS ONE. 2009;4:e7394. doi: 10.1371/journal.pone.0007394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee WH, Yue WW, Raush E, Totrov M, Abagyan R, Oppermann U, Marsden BD. Interactive JIMD articles using the iSee concept: turning a new page on structural biology data. J. Inherit. Metab. Dis. 2011;34:565–567. doi: 10.1007/s10545-011-9334-4. [DOI] [PubMed] [Google Scholar]
- 28.Abagyan R, Lee WH, Raush E, Budagyan L, Totrov M, Sundstrom M, Marsden BD. Disseminating structural genomics data to the public: from a data dump to an animated story. Trends Biochem. Sci. 2006;31:76–78. doi: 10.1016/j.tibs.2005.12.006. [DOI] [PubMed] [Google Scholar]
- 29.Lebon G, Warne T, Edwards PC, Bennett K, Langmead CJ, Leslie AGW, Tate CG. Agonist-bound adenosine A2A receptor structures reveal common features of GPCR activation. Nature. 2011;474:521–525. doi: 10.1038/nature10136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jaakola V-P, Griffith MT, Hanson MA, Cherezov V, Chien EYT, Lane JR, IJzerman AP, Stevens RC. The 2.6 Angstrom Crystal Structure of a Human A2A Adenosine Receptor Bound to an Antagonist. Science. 2008;322:1211–1217. doi: 10.1126/science.1164772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu F, Wu H, Katritch V, Han GW, Jacobson KA, Gao Z-G, Cherezov V, Stevens RC. Structure of an Agonist-Bound Human A2A Adenosine Receptor. Science. 2011;332:322–327. doi: 10.1126/science.1202793. [DOI] [PMC free article] [PubMed] [Google Scholar]