Abstract
The NucleaRDB is a Molecular Class-Specific Information System that collects, combines, validates and disseminates large amounts of heterogeneous data on nuclear hormone receptors. It contains both experimental and computationally derived data. The data and knowledge present in the NucleaRDB can be accessed using a number of different interactive and programmatic methods and query systems. A nuclear hormone receptor-specific PDF reader interface is available that can integrate the contents of the NucleaRDB with full-text scientific articles. The NucleaRDB is freely available at http://www.receptors.org/nucleardb.
INTRODUCTION
Nuclear receptors (NRs) are ligand-inducible transcription factors that regulate processes, such as homeostasis, differentiation, embryonic development and organ physiology. A total of 49 human NRs have been identified (1). Their ligands are lipophilic compounds such as steroids, thyroid hormone, vitamin D3 and retinoids (2). The endogenous ligands are not yet known for 30% of the NRs (3). As nuclear receptors are involved in almost all aspects of human physiology and are implicated in many important diseases including cancer, diabetes and osteoporosis, understanding of these receptors has major implications for human biology and for the development of new drug treatments. Nuclear receptors are targets for pharmaceutical industries with similar importance (4), as the G protein-coupled receptors (GPCRs), ion channels and kinases.
Due to the increasing amounts of experimental and computational data buried in numerous databases and scientific articles, the task of extracting, combining and validating this data is becoming an increasingly large hurdle for the individual scientist. Databases that revolve around a single protein family can help researchers in using all data needed for their research, while relieving them of the onerous tasks related to the retrieval of many data from different sources (5).
The NucleaRDB is a data source that holds many different data types (Table 1) in a well organized and easily accessible form (6). The data are validated, internally consistent and updated regularly. The NucleaRDB provides access to the data via various interfaces, which depending on the users’ needs, are suited either for automated access or interactive usage.
Table 1.
Contents of the NucleaRDB
| Proteins | 3764 |
| Families | 123 |
| Mutations | 1543 |
| Protein structures | 613 |
| Structure models | 3764 |
| Residues | 2 012 651 |
| Species | 339 |
DATA CONTENTS
Primary data
The NucleaRDB contains three different primary data types: sequences, structures and mutations. Sequences and structures were updated as described previously (7). Mutation data was obtained from the Nuclear Receptor Mutation Database (8) and fully integrated in the NucleaRDB. In addition, a large body of mutations was extracted from literature by the software package MuteXt (9).
Computational data
A large and diverse collection of computationally generated data are present in the NucleaRDB. Multiple sequence alignments (MSAs) form the heart of the system and allow users to easily transfer information between different proteins. MSAs are available for all families and subfamilies, and can be viewed using JalView (10) or can be directly downloaded in a number of formats. MSAs were created as described previously (7).
Correlated mutation analyses (CMA) can be used to identify groups of residues that mutate in tandem. Residues that show correlated mutation behavior are likely to be functionally related, and networks of those correlating residues indicate functional units (11). Correlation scores are available for all (sub-)families.
The entropy and variability for a position in a MSA can be an indicator of the evolutionary pressures exerted at that position (12). Entropy and variability scores are available in tabular form and via an interactive page displaying an integrated view via plots, tables and structure models.
In addition to the already large amount of structural information that is present in the NucleaRDB, homology models based on multiple template structures have been built for all NRs. All structure models were built using YASARA (13) and are available for download or can be viewed directly using Jmol (14).
INFORMATION RETRIEVAL
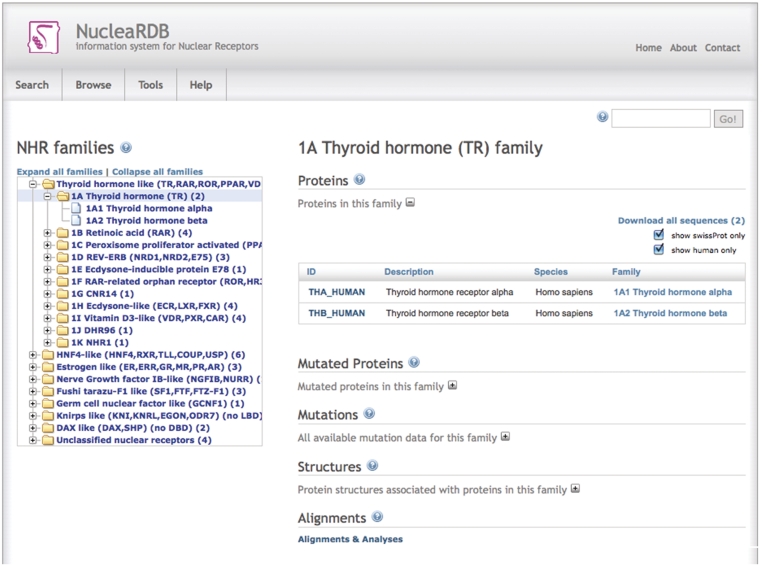
All data in the NucleaRDB web interface are extensively connected, allowing for easy navigation between different data types. The main way of accessing the NucleaRDB’s contents is via the hierarchical family tree. For each family, users can access the individual receptors, multiple sequence alignments (and all derived data and analyses such as correlation scores and protein distance networks), mutations, structures and models (Figure 1). All pages contain links to all related data and information. Extensive search facilities are available, allowing the search for proteins, sequences, structures, families and mutations using various search criteria and filters. A BLAST service is available that allows users to run their own sequences against the NucleaRDB.
Figure 1.
Screenshot of the NucleaRDB family page. The family tree is shown on the left with the thyroid hormone family expanded. On the right-hand side, the data for the selected family is shown.
All data types and search facilities are accessible from the web pages as well as from the web service endpoints, allowing users to write workflows or in-house software that uses the NucleaRDB.
Annotating scientific literature
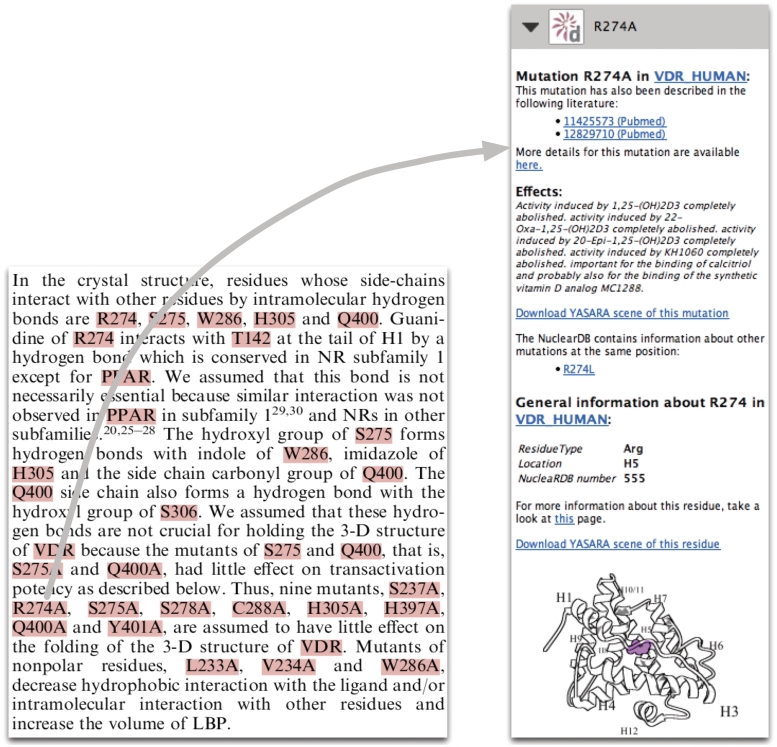
Utopia Documents (15,16) is a new PDF reader that offers unique opportunities to place information and knowledge in the context of scientific literature. We have integrated the NucleaRDB with the Utopia Documents PDF reader in such a way as to present to scientists, in a non-intrusive way, all NR-relevant data and information discussed in an article at hand. Annotations are provided for proteins, residues and mutations mentioned in the PDF. For each of these concepts the annotations contain carefully selected information, as well as pointers to relevant web pages and related scientific literature. An example is shown in Figure 2. The PDF reader presents the scientist, in a non-intrusive way, all relevant data and information related to the topics discussed in the article. This alleviates the troubles associated with navigating the many links between existing data and information available from the many articles in this field. The scientist neither struggles to get access to information related to topics within an article, nor is swamped by unnecessary information that still needs disambiguation; only data and information relevant to the topic of the article is made available.
Figure 2.
An impression of the Utopia Documents PDF reader interface to the NucleaRDB data. On the left-hand side a part of a scientific paper (17) is shown that is annotated by the NucleaRDB. Annotations are available for all the highlighted words. On the right-hand side an example of such an annotation (the mutation R274A) is displayed.
IMPLEMENTATION
The data in the NucleaRDB is stored in a PostgreSQL (www.postgresql.org) relational database. The web service interface is developed with the Apache CXF (cxf.apache.org) web services framework. We offer both Simple Object Access Protocol and Representational state transfer endpoints. The web interface is built using the Apache Wicket (wicket.apache.org) web application framework. The database is accessed via a Hibernate (www.hibernate.org) object-relational mapping layer. The server is running within Sun’s Glassfish (www.glassfish.org) application server.
CONCLUSION
The NucleaRDB provides researchers with a single point of access for nuclear receptor-related data. Not only does the NucleaRDB hold a large amount of information, it also provides a broad scope of tools and dissemination facilities, relieving scientist of many of the tasks that come with collecting, validating and integrating many diverse data.
FUNDING
BioRange program of the Netherlands Bioinformatics Centre (NBIC); BSIK grant through the Netherlands Genomics Initiative (NGI); EMBRACE project that is funded by the European Commission within its FP6 Programme, under the thematic area ‘Life sciences, genomics and biotechnology for health’ (contract number LHSG-CT-2004-512092); and TIPharma. Funding for open access charge: RUNMC.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank Maarten Hekkelman, Wilmar Teunissen and Tim teBeek for their support with computer science issues. We thank TIPharma for financial support.
REFERENCES
- 1.Robinson-Rechavi M, Carpentier AS, Duffraisse M, Laudet V. How many nuclear hormone receptors are there in the human genome? Trends Genet. 2001;17:554–556. doi: 10.1016/s0168-9525(01)02417-9. [DOI] [PubMed] [Google Scholar]
- 2.Mangelsdorf DJ, Thummel C, Beato M, Herrlich P, Schütz G, Umesono K, Blumberg B, Kastner P, Mark M, Chambon P, et al. The nuclear receptor superfamily: the second decade. Cell. 1995;83:835–839. doi: 10.1016/0092-8674(95)90199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kliewer SA, Lehmann JM, Willson TM. Orphan nuclear receptors: shifting endocrinology into reverse. Science. 1999;284:757–760. doi: 10.1126/science.284.5415.757. [DOI] [PubMed] [Google Scholar]
- 4.Hopkins AL, Groom CR. The druggable genome. Nat. Rev. Drug Discov. 2002;1:727–730. doi: 10.1038/nrd892. [DOI] [PubMed] [Google Scholar]
- 5.Folkertsma S, van Noort P, Van Durme J, Joosten H-J, Bettler E, Fleuren W, Oliveira L, Horn F, de Vlieg J, Vriend G. A family-based approach reveals the function of residues in the nuclear receptor ligand-binding domain. J. Mol. Biol. 2004;341:321–335. doi: 10.1016/j.jmb.2004.05.075. [DOI] [PubMed] [Google Scholar]
- 6.Horn F, Vriend G, Cohen FE. Collecting and harvesting biological data: the GPCRDB and NucleaRDB information systems. Nucleic Acids Res. 2001;29:346–349. doi: 10.1093/nar/29.1.346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vroling B, Sanders M, Baakman C, Borrmann A, Verhoeven S, Klomp J, Oliveira L, de Vlieg J, Vriend G. GPCRDB: information system for G protein-coupled receptors. Nucleic Acids Res. 2011;39:D309–D319. doi: 10.1093/nar/gkq1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Van Durme JJJ, Bettler E, Folkertsma S, Horn F, Vriend G. NRMD: Nuclear Receptor Mutation Database. Nucleic Acids Res. 2003;31:331–333. doi: 10.1093/nar/gkg122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Horn F, Lau AL, Cohen FE. Automated extraction of mutation data from the literature: application of MuteXt to G protein-coupled receptors and nuclear hormone receptors. Bioinformatics. 2004;20:557–568. doi: 10.1093/bioinformatics/btg449. [DOI] [PubMed] [Google Scholar]
- 10.Waterhouse AM, Procter JB, Martin DMA, Clamp M, Barton GJ. Jalview Version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Oliveira L, Paiva ACM, Vriend G. Correlated mutation analyses on very large sequence families. Chembiochem. 2002;3:1010–1017. doi: 10.1002/1439-7633(20021004)3:10<1010::AID-CBIC1010>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 12.Ye K, Lameijer E-WM, Beukers MW, Ijzerman AP. A two-entropies analysis to identify functional positions in the transmembrane region of class A G protein-coupled receptors. Proteins. 2006;63:1018–1030. doi: 10.1002/prot.20899. [DOI] [PubMed] [Google Scholar]
- 13.Krieger E, Joo K, Lee J, Lee J, Raman S, Thompson J, Tyka M, Baker D, Karplus K. Improving physical realism, stereochemistry, and side-chain accuracy in homology modeling: four approaches that performed well in CASP8. Proteins. 2009;77(Suppl. 9):114–122. doi: 10.1002/prot.22570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Herráez A. Biomolecules in the computer: Jmol to the rescue. Biochem. Mol. Biol. Educ. 2002;34:255–261. doi: 10.1002/bmb.2006.494034042644. [DOI] [PubMed] [Google Scholar]
- 15.Attwood TK, Kell DB, McDermott P, Marsh J, Pettifer SR, Thorne D. Calling international rescue: knowledge lost in literature and data landslide! Biochem. J. 2009;424:317–333. doi: 10.1042/BJ20091474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Attwood TK, Kell DB, McDermott P, Marsh J, Pettifer SR, Thorne D. Utopia documents: linking scholarly literature with research data. Bioinformatics. 2010;26:i568–i574. doi: 10.1093/bioinformatics/btq383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Choi M, Yamamoto K, Masuno H, Nakashima K, Taga T, Yamada S. Ligand recognition by the vitamin D receptor. Bioorg. Med. Chem. 2001;9:1721–1730. doi: 10.1016/s0968-0896(01)00060-8. [DOI] [PubMed] [Google Scholar]