Abstract
Proteins are flexible molecules that undergo structural changes to function. The Protein Data Bank contains multiple entries for identical proteins determined under different conditions, e.g. with and without a ligand molecule, which provides important information for understanding the structural changes related to protein functions. We gathered 839 protein structural pairs of ligand-free and ligand-bound states from monomeric or homo-dimeric proteins, and constructed the Protein Structural Change DataBase (PSCDB). In the database, we focused on whether the motions were coupled with ligand binding. As a result, the protein structural changes were classified into seven classes, i.e. coupled domain motion (59 structural changes), independent domain motion (70), coupled local motion (125), independent local motion (135), burying ligand motion (104), no significant motion (311) and other type motion (35). PSCDB provides lists of each class. On each entry page, users can view detailed information about the motion, accompanied by a morphing animation of the structural changes. PSCDB is available at http://idp1.force.cs.is.nagoya-u.ac.jp/pscdb/.
INTRODUCTION
Proteins are inherently flexible molecules, since their folded structures are mainly stabilized by non-covalent interactions. Although a static crystal structure provides an impression of a single rigid state, proteins exist in a range of structures. The Protein Data Bank (PDB) (1,2) contains a number of redundant entries for identical proteins with structures determined under different conditions, e.g. with and without a ligand molecule. These structures represent a range of variations in the native structures that are associated with their molecular functions (3–19). Recently, we collected the structural changes of proteins, represented by pairs of ligand-free and ligand-bound structures of identical proteins, to analyze whether the motions were coupled with ligand binding (20). The protein motions were classified into domain motions and local motions, depending on the size of the moving portions, and we identified the coupling with ligand binding by examining the relative positions between the ligand binding sites and the moving portions (20). Here, we have constructed the Protein Structural Change DataBase (PSCDB), which presents the detailed results of the analysis, i.e. the classifications of protein structural changes focusing on the coupling between protein motions and ligand binding. PSCDB will be an important resource to explore the essential motions for protein functions.
A few databases on protein structural changes, such as the database of macromolecular movements (21,22) and the non-redundant database of protein domain movements (23,24), are presently available. However, these databases have not focused on the causes of the structural changes. As a result, the databases contain various protein motions that are not necessarily essential to protein functions or artifacts due to crystal packing. Recently, Qi and Hayward (15) developed a database of protein motions induced by ligand binding, but they considered only domain motions in enzymes. PSCDB covers all kinds of protein motions, including not only domain motions of enzymes but also other types of motions seen in enzymes and non-enzymes.
METHODS
Data in PSCDB
Because the process of the dataset construction was previously described (20) in detail, we will briefly outline it here. We collected representative 839 structural changes of monomeric or homo-dimeric proteins, which are the pairs of the ligand-free and ligand-bound structures satisfying the following criteria: They are X-ray crystalline structures determined with higher than 3.0 Å resolution, and the sequence identity of the pair is >95%, with at least 90% coverage. The oligomeric states of the pair were identified according to PiQSi (Protein Quaternary Structure Investigation) (25). The representative pairs were selected according to the SCOP (structural classification of proteins) family (26), or based on clustering with 40% sequence identity. The representative pairs of ligand-free and ligand-bound structures of monomers and homo-dimers are a dominant part of the structural database: they occupy more than three quarters of the whole set of representative pairs including monomers and all kinds of oligomers.
Classification of protein motions
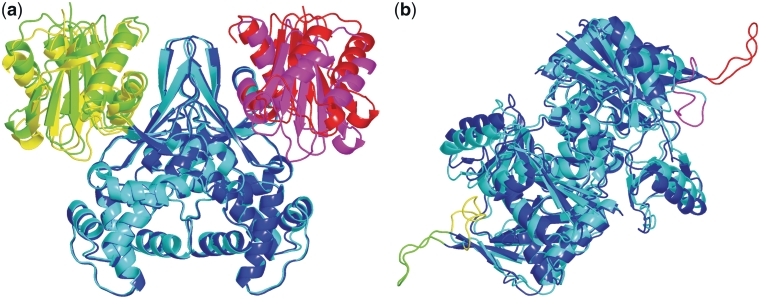
DynDom (27,28) was applied to the protein structures, regardless of monomeric or dimeric forms, and domain motions were detected. When more than two domains were identified, the protein structure was divided into the components of motion, each defined as two neighboring domains exhibiting a domain motion (Figure 1a). Consequently, a protein structure may have one or more components of motion, or simply a single domain exhibiting no domain motion. Next, the local motions were identified in each domain. A local motion was defined as that occurring in a local segment composed of more than five residues, with a displacement at least four times larger than the median for all residues. When more than one moving local segment was identified in a domain, the domain was divided into the components of motion, each composed of the core domain and a moving local segment (Figure 1b). Finally, each component of motion was assessed according to whether the motion was coupled with ligand-binding, by examining the relative positions of the ligand binding site and the moving portion. A component of motion was designated as ‘coupled’ with ligand-binding if both portions constituting the component of motion contacted the ligand molecule in the ligand-bound structure. Otherwise, it was classified as ‘independent’, or uncoupled. Consequently, the protein structural changes were classified as (i) ‘coupled domain motion’, (ii) ‘independent domain motion’, (iii) ‘coupled local motion’ and (iv) ‘independent local motion.’ In the classification of a structural change, the coexistence of domain and local motions in a protein was treated as domain motion, and the coexistence of coupled and independent motions was treated as coupled motion. The rest of the protein motions, other than the above four classes, were classified as follows. In the structural change with a small displacement [root-mean-square displacement (RMSD) <1.0 Å], we considered the protein structure to be significantly altered when the ligand molecule was buried inside of the protein core, i.e. the relative accessible surface area (31) of the ligand was >90%. We refer to this type of structural change as (v) ‘burying ligand motion’. We visually checked pocket shapes for ligands and confirmed that it would be impossible to bury the ligand without significant structural changes. The other motions with small RMSD values were termed (vi) ‘no significant motion’. The rest of the motions are (vii) ‘other type motion’, which are mostly outliers of the definitions of the domain or local motions, despite of large displacements (RMSD > 1.0 Å). They include ‘domain motion like’, ‘local motion like’ and ‘large plastic motion’ described in detail in the previous work (20). We mainly focused only on the coupled motions (classes 1 and 3) in our previous work (20), but PSCDB presents all motions.
Figure 1.
Components of motion in domain and local motions. (a) Superposition of glucokinase structures (PDBIDs: 1q18, 1sz2) (29) on the largest domains (blue/cyan). The three domains are colored green/yellow, blue/cyan and red/magenta. The structure was divided into two components of motion (green/yellow and blue/cyan, and red/magenta and blue/cyan). (b) Superposition of d-alanine:d-alanine ligase structures (PDBIDs: 2fb9, 2zdq) (30) on the core domains (blue/cyan). Two moving local segments (green/yellow and red/magenta) were identified in the protein motion. The structure was divided into two components of motion (green/yellow and blue/cyan, and red/magenta and blue/cyan).
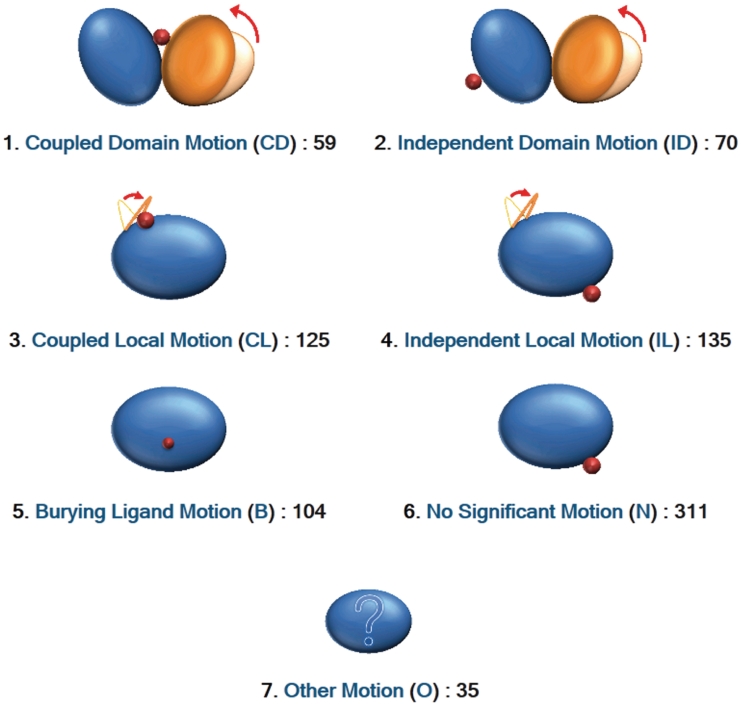
This classification is shown on the top-page of PSCDB, with the number of proteins classified into each class (Figure 2). Each class is represented by a code of one or two characters; for example, CD stands for ‘coupled domain motion’.
Figure 2.
Seven classes. The names, abbreviations, and numbers of protein structural changes of the seven classes are shown on the top-page of PSCDB, with the cartoon icons depicting each motion.
User interface
Users can view the list table of structural changes by clicking the icon of classification (Figure 2), or access the list table and the gallery of all entries by clicking the left box on the top-page. In the list table, the serial identifier, the PSCDB identifier (PSCID), is labeled on the individual structural change together with the protein name, the two PDBIDs (ligand-free and ligand-bound forms) and the ligand name(s). The six digit number shown in the right column in the list table indicates, from the first to the sixth digit, the number of components of motion for the coupled domain motion, the number for the independent domain motion, the number for the coupled local motion, the number for the independent local motion, the number of domains exhibiting the burying ligand motion, and the number of domains with ligand molecules situated on the protein surface, respectively. The last number is frequently related to the class of no significant motion. PSCDB also provides a search tool at the top-right corner of each page. Users can search for ‘protein name’ or ‘PDBID’.
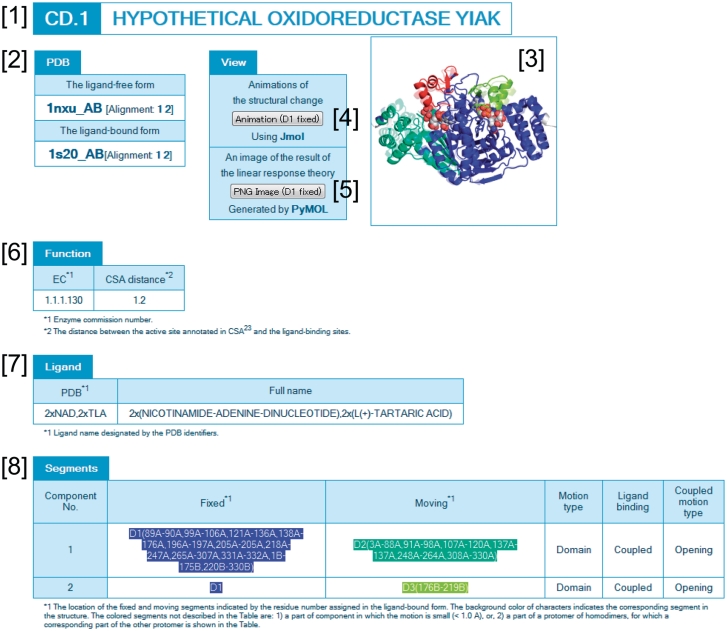
From the list table or the gallery, users can access the summary page of each protein structural change. As an example, the structural change of the hypothetical oxidoreductase YIAK (32) is shown in Figure 3. This protein contains two components of motion, which were both classified as coupled domain motion. The PSCID is thus CD.1 ([1] in Figure 3). The PDB structures representing the motion are provided in the ‘PDB’ box [2]. To visualize the motion, the three rigid domains (blue, green, and red) and the ligand molecules (spheres) are illustrated in the upper right panel [3]. By clicking the ‘animation’ button [4], morphing animation can be viewed through JMOL (33). The predicted domain motion using the linear response theory (20,34,35) can also be viewed by clicking the ‘PNG image’ button [5]. If a target protein is an enzyme, then the enzyme commission number and the information about the catalytic sites are shown in the ‘function’ box [6]. The bound-ligand molecule is provided in the ‘ligand’ box [7]. The details of the motions are summarized in the ‘segments’ box [8].
Figure 3.
An example of a web page: hypothetical oxidoreductase YIAK (32). The numbers indicate the PSCID and protein name [1], PDB box [2], protein and ligand structures [3], animation button [4], PNG image button [5], function box [6], ligand box [7] and segment box [8].
Future directions
In PSCDB, the structural changes are presented only for representative proteins of the SCOP families. The representative protein was selected as the pair of ligand-free and ligand-bound structures of an identical protein with the largest RMSD value in each protein family (20). However, the largest motion does not necessarily represent the dominant motion of the protein family. Therefore, we will examine all pairs of ligand-free and ligand-bound structures of an identical protein, and analyze the ensemble of protein motions in the protein family. We will also develop an automated pipeline to analyze and classify protein structural changes. Using this pipeline, PSCDB can be automatically updated when new PDB entries are deposited. A liaison to the BLAST (36) homology search program will also be available.
FUNDING
Funding for open access charge: Ministry of Education, Culture, Sports, Science and Technology of Japan.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors are grateful to Kiyotaka Misoo and Natsuko Hama for their contributions to the website design.
REFERENCES
- 1.Bernstein FC, Koetzle TF, Williams GJ, Meyer EF, Jr, Brice MD, Rodgers JR, Kennard O, Shimanouchi T, Tasumi M. The Protein Data Bank: a computer-based archival file for macromolecular structures. J. Mol. Biol. 1977;112:535–542. doi: 10.1016/s0022-2836(77)80200-3. [DOI] [PubMed] [Google Scholar]
- 2.Rose PW, Beran B, Bi C, Bluhm WF, Dimitropoulos D, Goodsell DS, Prlić A, Quesada M, Quinn GB, Westbrook JD, et al. The RCSB Protein Data Bank: redesigned web site and web services. Nucleic Acids Res. 2010;39:D392–D401. doi: 10.1093/nar/gkq1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tama F, Sanejouand YH. Conformational change of proteins arising from normal mode calculations. Protein Eng. 2001;14:1–6. doi: 10.1093/protein/14.1.1. [DOI] [PubMed] [Google Scholar]
- 4.Hammes GG. Multiple conformational changes in enzyme catalysis. Biochemistry. 2002;41:8221–8228. doi: 10.1021/bi0260839. [DOI] [PubMed] [Google Scholar]
- 5.Hayward S. Identification of specific interactions that drive ligand-induced closure in five enzymes with classic domain movements. J. Mol. Biol. 2004;339:1001–1021. doi: 10.1016/j.jmb.2004.04.004. [DOI] [PubMed] [Google Scholar]
- 6.Gutteridge A, Thornton J. Conformational change in substrate binding, catalysis and product release: an open and shut case? FEBS Lett. 2004;567:67–73. doi: 10.1016/j.febslet.2004.03.067. [DOI] [PubMed] [Google Scholar]
- 7.Gutteridge A, Thornton J. Conformational changes observed in enzyme crystal structures upon substrate binding. J. Mol. Biol. 2005;346:21–28. doi: 10.1016/j.jmb.2004.11.013. [DOI] [PubMed] [Google Scholar]
- 8.Alexandrov V, Lehnert U, Echols N, Milburn D, Engelman D, Gerstein M. Normal modes for predicting protein motions: a comprehensive database assessment and associated Web tool. Protein Sci. 2005;14:633–643. doi: 10.1110/ps.04882105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gunasekaran K, Nussinov R. How different are structurally flexible and rigid binding sites? Sequence and structural features discriminating proteins that do and do not undergo conformational change upon ligand binding. J. Mol. Biol. 2007;365:257–273. doi: 10.1016/j.jmb.2006.09.062. [DOI] [PubMed] [Google Scholar]
- 10.Keskin O. Binding induced conformational changes of proteins correlate with their intrinsic fluctuations: a case study of antibodies. BMC Struct. Biol. 2007;7:1–11. doi: 10.1186/1472-6807-7-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang L, Song G, Jernigan RL. How well can we understand large-scale proteins motions using normal modes of elastic network models. Biophys. J. 2007;93:920–929. doi: 10.1529/biophysj.106.095927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Koike R, Amemiya T, Ota M, Kidera A. Protein structural change upon ligand binding correlates with enzymatic reaction mechanism. J. Mol. Biol. 2008;379:397–401. doi: 10.1016/j.jmb.2008.04.019. [DOI] [PubMed] [Google Scholar]
- 13.Brylinski M, Skolnick J. What is the relationship between the global structures of apo and holo proteins? Proteins. 2008;70:363–377. doi: 10.1002/prot.21510. [DOI] [PubMed] [Google Scholar]
- 14.Dobbins SE, Lesk VI, Sternberg MJ. Insights into protein flexibility: the relationship between normal modes and conformational change upon protein–protein docking. Proc. Natl Acad. Sci. USA. 2008;105:10390–10395. doi: 10.1073/pnas.0802496105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Qi G, Hayward S. Database of ligand-induced domain movements in enzymes. BMC Struct. Biol. 2009;9:13. doi: 10.1186/1472-6807-9-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bakan A, Bahar I. The intrinsic dynamics of enzymes plays a dominant role in determining the structural changes induced upon inhibitor binding. Proc. Natl Acad. Sci. USA. 2009;106:14349–14354. doi: 10.1073/pnas.0904214106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boehr DD, Nussinov R, Wright PE. The role of dynamic conformational ensembles in biomolecular recognition. Nat. Chem. Biol. 2009;5:789–796. doi: 10.1038/nchembio.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grant BJ, Gorfe AA, McCammon JA. Large conformational changes in proteins: signaling and other functions. Curr. Opin. Struct. Biol. 2010;20:142–147. doi: 10.1016/j.sbi.2009.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sperandio O, Mouawad L, Pinto E, Villoutreix BO, Perahia D, Miteva MA. How to choose relevant multiple receptor conformations for virtual screening: a test case of Cdk2 and normal mode analysis. Eur. Biophys. J. 2010;39:1365–1372. doi: 10.1007/s00249-010-0592-0. [DOI] [PubMed] [Google Scholar]
- 20.Amemiya T, Koike R, Fuchigami S, Ikeguchi M, Kidera A. Classification and annotation of the relationship between protein structural change and ligand binding. J. Mol. Biol. 2011;408:568–584. doi: 10.1016/j.jmb.2011.02.058. [DOI] [PubMed] [Google Scholar]
- 21.Gerstein M, Krebs W. A database of macromolecular motions. Nucleic Acids Res. 1998;26:4280–4290. doi: 10.1093/nar/26.18.4280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Flores S, Echols N, Milburn D, Hespenheide B, Keateing K, Lu J, Wells S, Yu EZ, Gerstein M. The database of macromolecular motions: new features added at the decade mark. Nucleic Acids Res. 2006;34:D296–D301. doi: 10.1093/nar/gkj046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee R, Razaz M, Hayward S. The DynDom database of protein domain motions. Bioinformatics. 2003;19:1290–1291. doi: 10.1093/bioinformatics/btg137. [DOI] [PubMed] [Google Scholar]
- 24.Qi G, Lee R, Hayward S. A comprehensive and non-redundant database of protein domain movements. Bioinformatics. 2005;21:2832–2838. doi: 10.1093/bioinformatics/bti420. [DOI] [PubMed] [Google Scholar]
- 25.Levy ED. PiQSi: protein quaternary structure investigation. Structure. 2007;15:1364–1367. doi: 10.1016/j.str.2007.09.019. [DOI] [PubMed] [Google Scholar]
- 26.Andreeva A, Howorth D, Chandonia J-M, Brenner SE, Hubbard TJP, Chothia C, Murzin AG. Data growth and its impact on the SCOP database: new developments. Nucleic Acids Res. 2008;36:D419–D425. doi: 10.1093/nar/gkm993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hayward S, Berendsen HJC. Systematic analysis of domain motions in proteins from conformational change: new results on citrate synthase and T4 lysozyme. Proteins. 1998;30:144–154. [PubMed] [Google Scholar]
- 28.Hayward S, Lee RA. Improvements in the analysis of domain motions in proteins from conformational change: DynDom version 1.50. J. Mol. Graphics Model. 2002;21:181–183. doi: 10.1016/s1093-3263(02)00140-7. [DOI] [PubMed] [Google Scholar]
- 29.Lunin VV, Li Y, Schrag JD, Iannuzzi P, Cygler M, Matte A. Crystal structures of Escherichia coli ATP-dependent glucokinase and its complex with glucose. J. Bacteriol. 2004;186:6915–6927. doi: 10.1128/JB.186.20.6915-6927.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee JH, Na Y, Song HE, Kim D, Park B-H, Rho S-H, Im YJ, Kim MK, Kang GB, Lee D-S, et al. Crystal structure of the Apo form of D-Alanine: D-Alanine ligase (Ddl) from Thermus caldophilus: a basis for the substrate-induced conformational changes. Proteins. 2006;64:1078–1082. doi: 10.1002/prot.20927. [DOI] [PubMed] [Google Scholar]
- 31.Rose GD, Geselouitz AR, Lesser GJ, Lee RH, Zehfus MH. Hydrophobicity of amino acid residues in globular proteins. Science. 1985;30:834–838. doi: 10.1126/science.4023714. [DOI] [PubMed] [Google Scholar]
- 32.Forouhar F, Lee I, Benach J, Kulkarni K, Xiao R, Acton TB, Montelione GT, Tong L. A novel NAD-binding protein revealed by the crystal structure of 2,3-diketo-l-gulonate reductase (YiaK) J. Biol. Chem. 2004;279:13148–13155. doi: 10.1074/jbc.M313580200. [DOI] [PubMed] [Google Scholar]
- 33. Jmol: an open-source Java viewer for chemical structures in 3D. http://www.jmol.org/
- 34.Ikeguchi M, Ueno J, Sato M, Kidera A. Protein structural change upon ligand binding: linear response theory. Phys. Rev. Lett. 2005;94:1–4. doi: 10.1103/PhysRevLett.94.078102. [DOI] [PubMed] [Google Scholar]
- 35.Omori S, Fuchigami S, Ikeguchi M, Kidera A. Linear response theory in dihedral angle space for protein structural change upon ligand binding. J. Comput. Chem. 2009;30:2602–2608. doi: 10.1002/jcc.21269. [DOI] [PubMed] [Google Scholar]
- 36.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]