Abstract
ALFRED (http://alfred.med.yale.edu) is a free, web accessible, curated compilation of allele frequency data on DNA sequence polymorphisms in anthropologically defined human populations. Currently, ALFRED has allele frequency tables on over 663 400 polymorphic sites; 170 of them have frequency tables for more than 100 different population samples. In ALFRED, a population may have multiple samples with each ‘sample’ consisting of many individuals on which an allele frequency is based. There are 3566 population samples from 710 different populations with allele frequency tables on at least one polymorphism. Fifty of those population samples have allele frequency data for over 650 000 polymorphisms. Records also have active links to relevant resources (dbSNP, PharmGKB, OMIM, Ethnologue, etc.). The flexible search options and data display and download capabilities available through the web interface allow easy access to the large quantity of high-quality data in ALFRED.
INTRODUCTION
In this article, we are providing a detailed overview of ALFRED that has considerably evolved since the previous published descriptions >8 years ago (1–3). ALFRED is designed to be a resource for research and for education in diverse areas related to human genetic diversity. ALFRED's focus is on allele frequencies in diverse anthropologically defined populations. It is not a compendium of human DNA polymorphisms, but of allele frequencies of polymorphisms with an emphasis on those polymorphisms that have been studied in multiple populations. It is distinct from such databases as dbSNP (4,5), which is an uncurated catalog of sequence polymorphisms. We are not aware of any existing databases (private or public) other than ALFRED that attempts to meet the research needs of the broader human population genetics and molecular anthropology communities. There are many small and/or highly specialized databases. Applications such as FINDBase (6) (inherited disorders), STRbase (7) (forensic STRs), PharmGKB (8) (pharmacogenetic loci) and dbMHC (9) (HLA polymorphisms) are all excellent but specialized databases. All the data in ALFRED are considered to be in the public domain and available for the use in research and teaching.
Sources of data in ALFRED include the following: (i) data extracted from the published literature. Allele frequency data and related information are extracted from published papers located by ALFRED researchers and curators after routinely scanning the literature; (ii) data generated in the laboratories of K.K. and J.R. Kidd in the Department of Genetics at Yale, including extensive unpublished data; (iii) data submitted by collaborators or other researchers in electronic format; (iv) data in publicly available high-throughput SNP data sets such as the CEPH-HGDP data. Other high-throughput data are also being entered when possible to provide a single, more integrated resource. Intensive curation and data integrity checks are performed preceding any data upload into ALFRED.
Starting from our pre-existing database in 2000, we have progressively added more data (Table 1), improved the functionality of the web interface and elaborated the database structure. As of August 2011, there are 35 229 132 allele frequency tables (one population sample typed for one site) in ALFRED with additions ongoing on a regular basis.
Table 1.
The growth in contents summarized
| Records | 2000 | 2003 | 2006 | 2009 | Present |
|---|---|---|---|---|---|
| Frequency tables | 2865 | 11 505 | 39 298 | 1 149 836 | 35 230 146 |
| Polymorphisms | 180 | 698 | 1436 | ∼662 880 | 663 449 |
| Populations | 70 | 352 | 453 | 690 | 710 |
ALFRED continues to be supported by grants from the U.S. National Science Foundation to be an international resource for research and teaching.
DATABASE STRUCTURE AND CONTENT
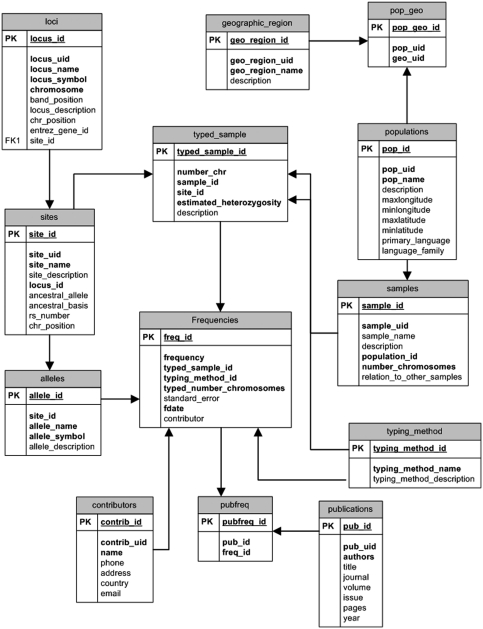
ALFRED has been implemented using relational database technology (Figure 1). All data are stored in an Oracle relational database management system. An individual polymorphism (or ‘Site’) is contained within a locus (‘Loci’ table) on the genome. Ethnic populations (‘Populations’ table) are organized by their geographic location (‘Geographic_Region’). Multiple samples (‘Samples’ table) may be drawn from a particular population. For such highly heterogeneous populations as African–American or European–American, special care is taken to delineate the specific geographic region of the population sample and to clearly distinguish among the multiple samples. The alleles at a site are in the ‘Alleles’ table. Because an allele frequency estimate is specific for a sample, the table ‘Typed_Sample’ bridges the tables Samples and Sites. The allele frequency values for a Typed_Sample are stored in the ‘Frequencies’ table with the associated typing method, which is detailed in the ‘Typing_Method’ table. All publication-related information is stored in a single ‘Publications’ table and intermediate tables are defined to link Publications to Frequencies, Samples, Sites and Loci. Links to other web sites are stored in the ‘URLs’ table. These links are associated with the Loci, Sites, Populations and Publication tables. All frequency records are linked to the contributor (Contributors table), which stores information about individuals who contribute the allele frequency data. Detailed descriptions of the individual tables (including their fields) are available from ‘Data structure’ (Table 2).
Figure 1.
Core Structure of ALFRED.
Table 2.
URLs to ALFRED pages mentioned in the text
| Page | URL |
|---|---|
| ALFRED home page | http://alfred.med.yale.edu |
| Table numbers | http://alfred.med.yale.edu/alfred/alfredsummary.asp |
| Data structure | http://alfred.med.yale.edu/alfred/table_list.asp |
| ALFRED wiki | http://alfred.med.yale.edu:8080/wiki/index.php/About_ALFRED |
| Feedback | http://alfred.med.yale.edu/alfred/feedback.asp |
| Downloads | http://alfred.med.yale.edu/alfred/alfredDataDownload.asp |
| Data submission | http://alfred.med.yale.edu/alfred/AboutALFRED.asp#datasubmission |
| Tour ALFRED | http://alfred.med.yale.edu/alfred/ALFREDtour-overview.asp |
| About ALFRED | http://alfred.med.yale.edu/alfred/AboutALFRED.asp |
| FAQ | http://alfred.med.yale.edu/alfred/alfredFaq.asp |
| ALFRED flyer | http://alfred.med.yale.edu/alfred/flyer/ALFREDFlyer.pdf |
ACCESS TO THE DATA IN ALFRED
Specific information in ALFRED can be accessed in multiple ways through the web. Users familiar with the Google search engine may search for an rs number, gene symbol, population or ALFRED UID by simply concatenating the string ALFRED to the search term. For example, a search term like ‘ALFRED E_rs1 587 264_10’ or ‘ALFRED SI001677V’ will list the URL link to ALFRED's ‘Polymorphism Information’ page for the rs number rs1 587 264. E_rs1 587 264_10 is the TaqMan assay in the Applied Biosystems catalog used to obtain the allele frequency; SI001677V is the UID of the record in ALFRED. A simple rs number or gene name would work as well. Similarly, concatenating a population name or a specific population UID may bring up the link to the corresponding ‘Population Information’ page. This use of Google requires prior knowledge of one of the terms in ALFRED and does not always retrieve the result desired but is very quick when it does work.
Usually, specific information in ALFRED will be accessed through the ALFRED web interface, which offers multiple options. The ALFRED web site also allows direct access to a specific record using the keyword search function available on the ALFRED home page. Users have the option of selecting the type of search, ‘Any part of’ or ‘Begins with’ and the table that should be searched. The option ‘Any part of’ considers ALFRED names that contain the entered string of characters anywhere, while ‘Begins with’ only considers ALFRED names that begin with the entered string of characters. In addition, the search can be restricted to the database table to be searched. The resulting output is a comprehensive table of the different occurrences of the search term, the database table in which it occurs and a link to navigate to the corresponding description page. Users looking for a specific SNP with dbSNP refSNP Identifier (rs number), gene symbol or a population can take advantage of this search option.
A more generalized method for searching ALFRED without specific prior criteria is by following the two options under the tabbed menu item Search: Loci and Population. The returned results are organized as follows. Loci are organized both in genomic order by chromosome and molecular position as well as in alphabetic order. Following either of the options, selecting a locus will then bring the user to the specific Locus Information page. Each locus record is annotated with alternate names (synonyms), chromosomal position, a valid HUGO Nomenclature Committee locus symbol and links to external databases such as Entrez Gene, UniGene, OMIM, PharmGKB and Genopedia (HuGE Navigator). Genetic polymorphisms and haplotypes ordered by chromosomal position in the selected locus are displayed in a table. For example, see (http://alfred.med.yale.edu/alfred/recordinfo.asp?UNID=LO000422I). A polymorphism or haplotype can be selected to navigate to the Polymorphism Information page. Each polymorphism record is annotated with dbSNP rs number, alternate names (synonyms), ancestral allele and links to external databases such as dbSNP and PharmGKB for expanded molecular information. For example, see (http://alfred.med.yale.edu/alfred/recordinfo.asp?UNID=SI000002C). Populations are organized by geographic regions and selecting a population will bring the user to the corresponding Population Information page. Each population record is annotated with alternate names (synonyms), linguistic, geographical location information and links to external databases such as Ethnologue Language and Map Projects for additional information. Active links to other databases provided from ALFRED's populations, loci, and sites information pages facilitate easy retrieval of additional information. For example, see (http://alfred.med.yale.edu/alfred/recordinfo.asp?UNID=PO000036J).
Population samples are organized by populations and annotated with sample information such as sample size and relation to other samples. The wiki implementation for ALFRED ‘ALFRED Wiki’ (Table 2) allows users to interact with ALFRED curators and get involved in annotating ALFRED populations. ALFRED curators are responsible for comparing between different wiki update versions and adding relevant information to the population descriptions in ALFRED. We invite ALFRED users to participate in this effort of population annotation. Users are required to create an account and log in to be able to edit the ALFRED wiki pages. Contact us using our feedback function on the ALFRED home page to create an account.
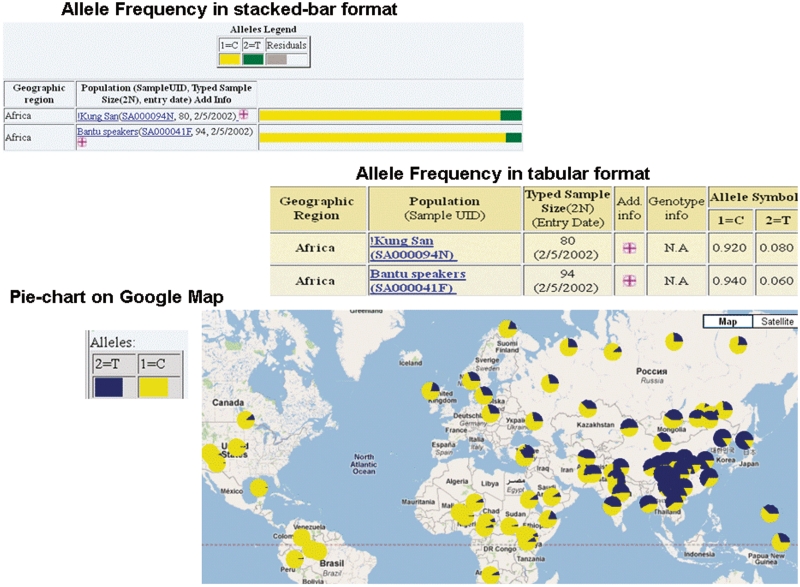
Allele frequency records are accessed from the corresponding site (Polymorphism Information) page. Display formats available are graphical, tabular and pie-chart on Google Map (Figure 2). The graphically stacked-bar format offers a quick visual display of the frequency variation among populations (http://alfred.med.yale.edu/alfred/mvograph.asp?siteuid=SI001272M). Each allele frequency record displayed is linked to the population sample information, polymorphism information, typing method and the publication the frequency was extracted from. Most publication entries are linked to PubMed for complete citation and possible links to the full publication. For diallelic polymorphisms, the web page also provides a table with the calculated Fst, average heterozygosity (measures of genetic variation) and number of populations with data available in ALFRED for the selected site. The graphical stacked-bar format and the pie-chart on Google Map offer quick visual displays of the frequency variation among populations (http://alfred.med.yale.edu/alfred/mvograph.asp?siteuid=SI014485W). On the other hand, the tabular format gives the frequency values and related information, which can be used in analyses (see also Downloads). (http://alfred.med.yale.edu/alfred/SiteTable1A_working.asp?siteuid=SI001272M).
Figure 2.
Different allele frequency display formats for rs2 066 701 of ADHIB gene.
Every record in ALFRED has a unique identifier (UID) that can be the basis of a search; the UID search option is under the Search tab. While this search option is not used very often, it can be very effective. The UIDs are a text string consisting of three parts: for example, LO000423J is the UID for the locus ADH4 (the prefix ‘LO’ indicates the UID refers to a locus, the suffix J is the Check Character and 000423 is a number generated by the system when the record is created). Searching ALFRED with an UID (Site, Population, Locus or Sample) will bring the user to the corresponding ‘Information’ page. We have found it very useful in human interactions to have the human interpretable prefixes as part of the UID schema. Similarly, the check character helps prevent a false retrieval that could result from a numeric typo.The SNP sets page under the ‘Search’ tab facilitates user access to defined SNP sets published for ancestry inference and forensic individual identification. The markers in each of these SNP sets are annotated with relevant information including the locus name, rs number, Fst, average heterozygosity and the number of populations for which there are data available in ALFRED. The page listing all of the SNPs in a set has options for sorting by each of those values. Each record in a set links out to locus description page, site description page and to the ‘Google Map’ with pie-chart distribution of the allele frequencies (see also Downloads).
DATA DOWNLOAD FORMATS
Several options are available for retrieving data from ALFRED for various analyses by the user. Every individual Polymorphism Information page allows allele frequency download in several formats including both tab-delimited text and the input file format for the population genetics software package ‘Arlequin’. These download options yield data comparable to the tabular display format; each record gives the population name, the sampleUID, and the frequencies of the two alleles. The download will include a record for every sample for which there are data. The field ‘entryDate’ in the file can be used to distinguish between allele frequencies on the same sample. A complete allele frequency data dump can be obtained by downloading the ‘alfredFreq.zip’ or ‘alfredFreqByChrom.zip’ zipped files. The tables are in text format (tab-delimited), which can easily be parsed and opened in any text editor or MS Excel spreadsheet. Similarly, all the sites and related information from the Sites, Loci and Allele tables can be obtained by downloading ‘alfredPolymorphisms.zip’, while the Populations table is in ‘alfredPops.zip’. All these files can be downloaded from ‘Downloads’ (Table 2). Allele frequency tables for selected SNP sets can be downloaded from the ‘Downloads’ page as well. As new interesting SNP sets are added to ALFRED the data will be made available for download. The zip files are updated on every Friday.
LINKING TO ALFRED
In addition to the files listed above, two mapping tables can be downloaded: one maps ALFRED UID for loci to Entrez Gene Id (ALFREDGeneInfo.csv), and the other maps ALFRED UID for sites to dbSNP rs number (ALFREDVariantInfo.csv). Very often related resources on the web are interlinked by providing URLs to and from relevant pages. These mapping tables will facilitate easy creation of URLs to ALFRED. Based on UIDs, anyone can create URLs to locus and site description pages in ALFRED using the following format: http://alfred.med.yale.edu/alfred/recordinfo.asp?UNID= <UID> (where <UID> will be replaced by the actual UID value). The above-mentioned two mapping tables have facilitated reciprocal URLs from PharmGKB, and CDC’s HuGE Navigator (10). In addition, reciprocal URLs from the dbSNP rs number page to ALFRED’s Polymorphism Information page are maintained by periodically submitting a dbSNP-specified XML file.
HIGHLIGHTS OF DATA IN ALFRED
Over the years, there have been several interesting allele frequency additions to ALFRED.
High-throughput data sets in ALFRED worth mentioning are:
Over 350 autosomal short tandem repeat polymorphisms typed on the CEPH-HGDP human diversity panel, which includes 51 worldwide populations. These polymorphisms are located throughout the genome (11);
Over 11 555 SNPs typed on 14 populations (12);
Over 650 000 common SNPs typed by Illumina technology (650Ypanel) on the CEPH-HGDP panel of 51 populations (13). In addition, 876 markers from this set typed on 46 Kidd Lab population samples are in ALFRED; and
Over 2800 SNPs typed on the CEPH-HGDP panel and an additional two Indian populations (total of 55 samples) (14).
Other smaller but interesting data additions to ALFRED (allele frequency tables for these sets are available from the ‘Downloads’ page):
Thirty-four-plex assay markers data on the CEPH-HGDP panel from Phillips et al. (15). In addition, for these markers data typed on 46 Kidd Lab populations are in ALFRED bringing the total to 98 population samples;
Fifty-two ‘SNPforID’ markers typed on 16 population samples from Sanchez et al. (16). Several of these markers have subsequently been typed on additional populations and data will be added to ALFRED;
‘LowFst’ markers of forensic interest typed on the Kidd Lab population panel (17, 18);
One hundred and twenty-eight ancestry informative markers typed on 73 Kidd Lab populations (19);
Various interesting polymorphisms associated with human traits (20, 21);
Polymorphisms associated with ‘lactase persistence’ (22); and
TAS2R16 gene-coding polymorphisms typed on the HGDP-CEPH panel (23) and Kidd Lab populations (24).
USER INVOLVEMENT
We encourage users to communicate with us on the interface or any data contained in ALFRED using the ‘Feedback’ page. Allele frequency data can be submitted to us electronically by following the directions in the guidelines for ‘Data submission’ (Table 2).
Comprehensive and up-to-date documentation of the contents and navigation tips can be obtained from ‘Tour ALFRED’, ‘About ALFRED’, ‘ALFRED FAQ’ and ‘ALFRED flyer’ (Table 2).
FUTURE DIRECTIONS
The number of records in ALFRED will continue to grow as allele frequency data for new population samples and SNPs are made available. During the coming month's, data from the Illumina 650Y panel will be entered for several additional populations. Also, data download options of a user-selected set of SNPs and populations will be implemented. We also hope to enhance the didactic value of the database. On these and other directions for the future, we welcome comments and suggestions toward better meeting needs of the community.
FUNDING
U.S. National Science Foundation (grant BCS0938633). Funding for open access charge: U.S. National Science Foundation (grant BCS0938633).
Conflict of interest statement. None declared.
REFERENCES
- 1.Cheung KH, Osier MV, Kidd JR, Pakstis AJ, Miller PL, Kidd KK. ALFRED: an allele frequency database for diverse populations and DNA polymorphisms. Nucleic Acids Res. 2000;28:361–363. doi: 10.1093/nar/28.1.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Osier MV, Cheung KH, Kidd JR, Pakstis AJ, Miller PL, Kidd KK. ALFRED: an allele frequency database for Anthropology. Am. J. Phys. Anthropol. 2002;119:77–83. doi: 10.1002/ajpa.10094. [DOI] [PubMed] [Google Scholar]
- 3.Rajeevan H, Osier MV, Cheung KH, Deng H, Druskin L, Heinzen R, Kidd JR, Stein S, Pakstis AJ, Tosches NP, et al. ALFRED – the ALlele FREquency Database (Update) Nucleic Acids Res. 2003;31:270–271. doi: 10.1093/nar/gkg043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sherry ST, Ward MH, Kholodov M, Baker J, Phan L, Smigielski EM, Sirotkin K. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sayers EW, Barrett T, Benson DA, Bolton E, Bryant SH, Canese K, Chetvernin V, Church DM, Dicuccio M, Federhen S, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2010;38:D5–D16. doi: 10.1093/nar/gkp967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van Baal S, Kaimakis P, Phommarinh M, Koumbi D, Cuppens H, Riccardino F, Macek M, Jr, Scriver CR, Patrinos GP. FINDbase: a relational database recording frequencies of genetic defects leading to inherited disorders worldwide. Nucleic Acids Res. 2007;35:D690–D695. doi: 10.1093/nar/gkl934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ruitberg CM, Reeder DJ, Butler JM. STRBase: a short tandem repeat DNA database for the human identity testing community. Nucleic Acid Res. 2001;29:320–322. doi: 10.1093/nar/29.1.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hewett M, Oliver DE, Rubin DL, Easton KL, Stuart JM, Altman RB, Klein TE. PharmGKB: the Pharmacogenetics Knowledge Base. Nucleic Acids Res. 2001;30:163–165. doi: 10.1093/nar/30.1.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gourraud PA, Feolo M, Hoffman D, Helmberg W, Cambon-Thomsen A. The dbMHC microsatellite portal: a public resource for the storage and display of MHC microsatellite information. Tissue Antigens. 2006;67:395–401. doi: 10.1111/j.1399-0039.2006.00600.x. [DOI] [PubMed] [Google Scholar]
- 10.Yu W, Gwinn M, Clyne M, Yesupriya A, Khoury MJ. A navigator for human genome epidemiology. Nat. Genet. 2008;40:124–125. doi: 10.1038/ng0208-124. [DOI] [PubMed] [Google Scholar]
- 11.Rosenberg NA, Pritchard JK, Weber JL, Cann HM, Kidd KK, Zhivotovsky LA, Feldman MW. Genetic structure of human populations. Science. 2002;298:2381–2385. doi: 10.1126/science.1078311. [DOI] [PubMed] [Google Scholar]
- 12.Shriver MD, Mei R, Parra EJ, Sonpar V, Halder I, Tishkoff SA, Schurr TG, Zhadanov SI, Osipova LP, Brutsaert TD, et al. Large-scale SNP analysis reveals clustered and continuous patterns of human genetic variation. Hum. Genomics. 2005;2:81–89. doi: 10.1186/1479-7364-2-2-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li JZ, Absher DM, Tang H, Southwick AM, Casto AM, Ramachandran S, Cann HM, Barsh GS, Feldman M, Cavalli-Sforza LL, et al. Worldwide human relationships inferred from genome-wide patterns of variation. Science. 2008;319:1100–1104. doi: 10.1126/science.1153717. [DOI] [PubMed] [Google Scholar]
- 14.Pemberton TJ, Jakobsson M, Conrad DF, Coop G, Wall JD, Pritchard JK, Patel PI, Rosenberg NA. Using population mixtures to optimize the utility of genomic databases: linkage disequilibrium and association study design in India. Ann. Hum. Genet. 2008;72:535–546. doi: 10.1111/j.1469-1809.2008.00457.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Phillips C, Salas A, Sánchez JJ, Fondevila M, Gómez-Tato A, Álvarez-Dios J, Calaza M, de Cal CM, Ballard D, Lareu MV, et al. The SNPforID Consortium. Inferring ancestral origin using a single multiplex assay of ancestry-informative marker SNPs. Forensic Sci. Int. Genet. 2007;1:273–280. doi: 10.1016/j.fsigen.2007.06.008. [DOI] [PubMed] [Google Scholar]
- 16.Sanchez JJ, Phillips C, Borsting C, Balogh K, Bogus M, Fondevila M, Harrison CD, Musgrave-Brown E, Salas A, Syndercombe-Court D, et al. A multiplex assay with 52 single nucleotide polymorphisms for human identification. Electrophoresis. 2006;27:1713–1724. doi: 10.1002/elps.200500671. [DOI] [PubMed] [Google Scholar]
- 17.Kidd KK, Pakstis AJ, Speed WC, Grigorenko EL, Kajuna SL, Karoma NJ, Kungulilo S, Kim JJ, Lu RB, Odunsi A, et al. Developing a SNP panel for forensic identification of individuals. Forensic Sci. Int. 2006;164:20–32. doi: 10.1016/j.forsciint.2005.11.017. [DOI] [PubMed] [Google Scholar]
- 18.Pakstis AJ, Speed WC, Kidd JR, Kidd KK. Candidate SNPs for a Universal Individual Identification Panel. Hum. Genet. 2007;121:305–317. doi: 10.1007/s00439-007-0342-2. [DOI] [PubMed] [Google Scholar]
- 19.Kosoy R, Nassir R, Tian C, White PA, Butler LM, Silva G, Kittles R, Alarcon-Riquelme ME, Gregersen PK, Belmont JW, et al. Ancestry informative marker sets for determining continental origin and admixture proportions in common populations in America. Hum. Mutat. 2009;30:69–78. doi: 10.1002/humu.20822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yoshiura KI, Kinoshita A, Ishida T, Ninokata A, Ishikawa T, Kaname T, Bannai M, Tokunaga K, Sonoda S, Komaki R, et al. A SNP in the ABCC11 gene is the determinant of human earwax type. Nat. Genet. 2006;38:324–330. doi: 10.1038/ng1733. [DOI] [PubMed] [Google Scholar]
- 21.Nadkarni NA, Weale ME, von Schantz M, Thomas MG. Evolution of a length polymorphism in the human PER3 gene, a component of the circadian system. J. Biol. Rhythms. 2005;20:490–499. doi: 10.1177/0748730405281332. [DOI] [PubMed] [Google Scholar]
- 22.Coelho M, Luiselli D, Bertorelle G, Lopes AI, Seixas S, Destro-Bisol G, Rocha J. Microsatellite variation and evolution of human lactase persistence. Hum. Genet. 2005;117:329–339. doi: 10.1007/s00439-005-1322-z. [DOI] [PubMed] [Google Scholar]
- 23.Soranzo N, Bufe B, Sabeti PC, Wilson JF, Weale ME, Marguerie R, Meyerhof W, Goldstein DB. Positive selection on a high-sensitivity allele of the human bitter-taste receptor TAS2R16. Curr. Biol. 2005;15:1257–1265. doi: 10.1016/j.cub.2005.06.042. [DOI] [PubMed] [Google Scholar]
- 24.Li H, Pakstis AJ, Kidd JR, Kidd KK. Selection on the human bitter taste gene, TAS2R16, in Eurasian populations. Hum. Biol. 2011;83:363–377. doi: 10.3378/027.083.0303. [DOI] [PubMed] [Google Scholar]