Abstract
In the post-genomic era, most components of a cell are known and they can be quantified by large-scale functional genomics approaches. However, genome annotation is the bottleneck that hampers our understanding of living cells and organisms. Up-to-date functional annotation is of special importance for model organisms that provide a frame of reference for studies with other relevant organisms. We have generated a Wiki-type database for the Gram-positive model bacterium Bacillus subtilis, SubtiWiki (http://subtiwiki.uni-goettingen.de/). This Wiki is centered around the individual genes and gene products of B. subtilis and provides information on each aspect of gene function and expression as well as protein activity and its control. SubtiWiki is accompanied by two companion databases SubtiPathways and SubtInteract that provide graphical representations of B. subtilis metabolism and its regulation and of protein–protein interactions, respectively. The diagrams of both databases are easily navigatable using the popular Google maps API, and they are extensively linked with the SubtiWiki gene pages. Moreover, each gene/gene product was assigned to one or more functional categories and transcription factor regulons. Pages for the specific categories and regulons provide a rapid overview of functionally related genes/proteins. Today, SubtiWiki can be regarded as one of the most complete inventories of knowledge on a living organism in one single resource.
INTRODUCTION
The investigation of model organisms is a key element in the development of our understanding of biological processes. The availability of information on genes, proteins and cellular processes in this handful of organisms is essential not only for the better understanding of these organisms, but also as a benchmark to study related organisms that may be more relevant with respect to medical or biotechnological aspects.
Bacillus subtilis is a well-characterized model organism for Gram-positive bacteria that include important pathogens such as Bacillus anthracis, Listeria monocytogenes or Staphylococcus aureus, as well as biotechnologically important species such as Bacillus licheniformis and the lactic acid bacteria. With the increasing amount of knowledge gained by functional genomics studies, B. subtilis is today one of the most advanced model organisms for systems biology approaches and is even considered as a potential host for synthetic modules. Due to the considerable importance of B. subtilis, a comprehensive and up-to-date source of information on the genes and proteins, their regulation, interactions and associated pathways is required, allowing the busy researcher to keep pace with the continuously accumulating data and knowledge.
In parallel to the first genome sequencing project (1), the SubtiList database was established and became very popular in the scientific community working with Gram-positive bacteria (2). Unfortunately, it has not been updated since 2005 and is no longer available since 2010. Recently, SubtiList was integrated into a larger database GenoList that aims at providing comprehensive information on multiple bacterial genomes (http://genodb.pasteur.fr/cgi-bin/WebObjects/GenoList.woa/10/wa/goToTaxoRank?level=Bacillus%20subtilis%20168). In order to provide the community with up-to-date information, we created the Wiki-based data source SubtiWiki (3,4). The decision to make use of a Wiki was based on the experience that a database run by a single institution might no longer be updated if the focus of that institution changes. To the Wiki, each qualified scientist can contribute, thus the combined knowledge of the Bacillus community can be collected and made accessible to anyone interested in any specific aspect related to B. subtilis and other Gram-positive bacteria.
When SubtiWiki was created, it was the idea to collect all the information related to a gene/protein on a single page and provide this information with links to the relevant evidence (i.e. the PubMed entries). Moreover, the possibility to create internal links was used to facilitate the discovery of relations between different genes, proteins or RNAs (3). With the ongoing use and development of SubtiWiki it became obvious that it is well suited as a platform for different types of information. In a first attempt, graphical models of B. subtilis metabolic and regulatory pathways were created using CellDesigner in the Systems biology markup language (SBML) (5) and linked to the SubtiWiki pages. This resulted in a suite of graphical representations that is called SubtiPathways (4).
The access to the currently available knowledge in an appropriate form is pivotal to the interpretation of data from genome-scale experiments that are relevant to systems and synthetic biology. Therefore, SubtiWiki was further developed to support data mining approaches by providing a functional classification of the gene products of B. subtilis and a comprehensive collection of transcription factor regulons. These data sets are available in formats that are directly suitable for bioinformatic applications.
The genomic era and the initial phase of systems biology have provided us with lists of components that make up cells. Today, we can identify and quantify almost all molecules in a living cell, including proteins, RNA species and metabolites. However, only the interactions between these molecules make real life out of the components. In the past few years, these interactions came more and more into the focus of scientific research and also the B. subtilis community spent a lot of efforts to elucidate protein–protein interactions (6–11). In its initial stage, SubtiWiki was focused on the individual components of the cell, and their interactions were just one aspect among many others. Given the great importance of protein–protein interactions we decided to accompany SubtiWiki by yet another project, SubtInteract, which provides information on protein–protein interactions and that is again closely interconnected with SubtiWiki.
Due to the great importance of B. subtilis as a model organism, additional initiatives have started to provide better functional annotation for this organism. Specifically, BsubCyc is part of the BioCyc collection of databases (12). According to the authors, BsubCyc is moderately curated. Moreover, a second Wiki for B. subtilis, SubtilisWiki, has recently been set up which is still in its initial stage.
In this work, we describe the current state of SubtiWiki with special emphasis on the new features: (i) the functional classification of the B. subtilis gene products, (ii) the compilation and implementation of transcription factor regulons and (iii) SubtInteract for the visualization of protein–protein interaction networks in B. subtilis.
The key feature of SubtiWiki: the pages for individual genes and proteins
In SubtiWiki, there is an individual page for each gene that provides all the information on both the gene and its product, usually a protein, sometimes an RNA. In the original version of SubtiWiki, these pages could be retrieved by the gene designation. To take account of changing gene names and the fact that not every user might be familiar with these names, the pages can now also be accessed using a fixed identifier, the so-called locus tag that was given to each gene or genetic feature when the B. subtilis genome was annotated (1, 13).
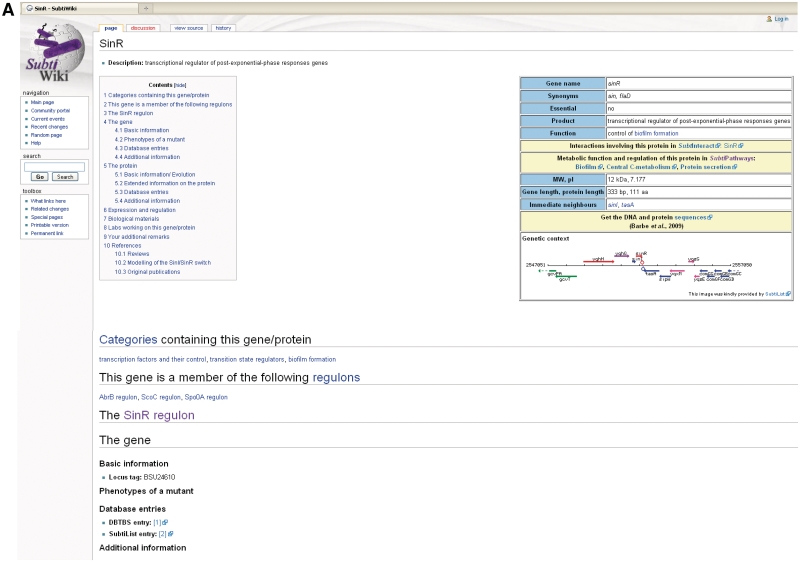
An example of a SubtiWiki gene page is shown in Figure 1. At the top of each page, the contents and a table with the most important information are shown. This table covers the gene name and synonym designations, the gene product and its function, the molecular weight and isoelectric point of the protein, the gene and protein lengths and the names of the adjacent genes. Moreover, the table contains links to the relevant representations of protein–protein interactions and to the pathway diagrams. Finally, the DNA and protein sequences can be downloaded and a diagram of the genomic context is shown.
Figure 1.
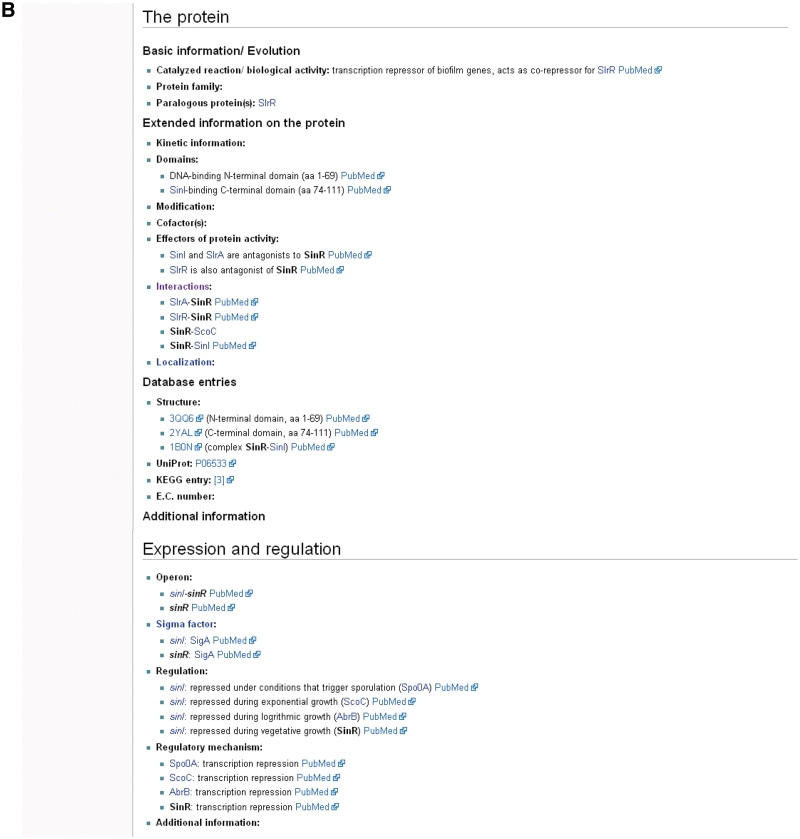
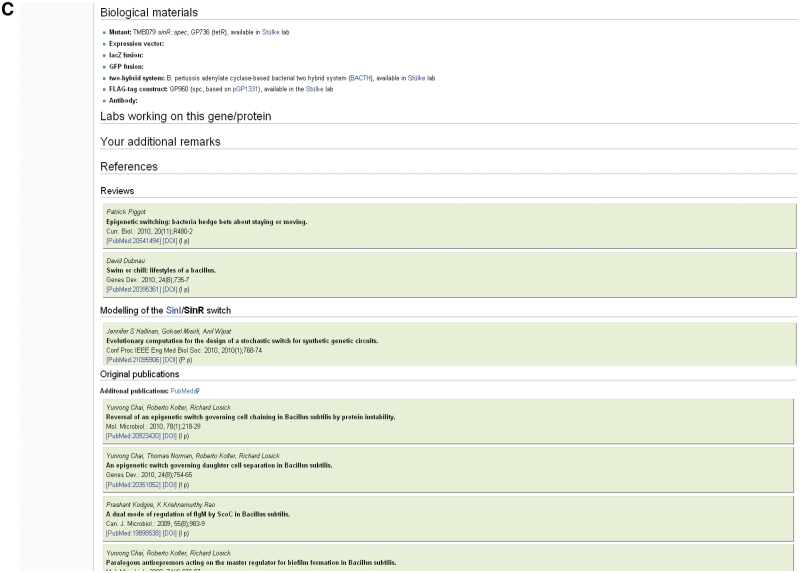
The sinR gene page in SubtiWiki. Pages for each gene of B. subtilis exist in SubtiWiki. Background information on the gene (A) and protein (B) are given, as well as links to SubtiPathways and SubtInteract. The bottom part of the pages displays community information and references (C) (Note that some publications have temporarily been left out for the purpose of this figure).
Below the table, the functional categories and regulons for the gene/protein are shown (see below). This allows immediate access to all related genes or proteins that are members of the same category or regulon. In the case of transcription regulators, a link to the page dedicated to the regulon controlled by the regulatory protein is provided.
The next section contains the information about the gene. It covers basic information as the unique identifier (locus tag), phenotypes of a mutant and gene-specific database entries. The largest section of each page is devoted to the encoded protein. It provides information on the biological activity of the protein, the protein family and possible paralogs. Moreover, kinetic data (if available), as well as information on protein domains, modifications, co-factors, effectors, interactions and the localization are presented. As for the gene section, the section for the protein is concluded with database links, such as structure databases (e. g. PDB), Uniprot or KEGG entries, as well as the E.C. numbers.
The following section of the page provides information on the expression of the gene and its regulation. This includes the operon structure, the sigma factor, transcription factors and their mode of action. At the bottom of the page, there is some community related information (biological materials, labs working on the gene/protein), as well as a collection of references on all aspects of the gene/protein.
New features of SubtiWiki: the functional classification of the B. subtilis gene products and a comprehensive collection of transcription factor regulons
The current version of SubtiWiki describes the relatedness between the individual genes also by assigning common functional and regulatory properties. To this end, we first established a systematic functional classification of the B. subtilis gene products which is described in detail in the next paragraph. Second, we compiled the transcription factor regulons by collecting and manually curating information from DBTBS (14), a database of published transcriptional regulation events in B. subtilis. In addition, information on regulation was manually extracted from the recent literature. With respect to the target genes of individual transcription factors, we consulted scientists with expertise in specific fields of B. subtilis transcriptional regulation.
A first functional classification scheme was applied to all protein coding genes of B. subtilis (15) after completion of the genome sequence (1) and implemented into the SubtiList database (2). This scheme was adapted from a classification originally devised for Escherichia coli (16). However, in recent years it became apparent that the functional classification from SubtiList was no longer adequate because, (i) functions had been assigned to many ‘y-genes’ since the last update in 2002, (ii) the new B. subtilis genome sequence and annotation (13) introduced around 200 new genes, either newly annotated or resulting from fusions or fissions of previously existing genes and (iii) the classification scheme allowed each gene to be assigned only to a single annotation term. As for the majority of the gene products, the physiological role cannot be sufficiently described by a single functional category (17), more advanced functional classification schemes for bacterial species have been developed that permit the assignment of multiple categories to one gene product and also possess a higher specificity through several layers of subcategories (18,19). Consequently, a new functional classification for the B. subtilis gene products has been devised and implemented in SubtiWiki. This classification is organized in a hierarchical, tree-like structure with six main categories that are subdivided in up to four levels of increasing specificity. Five of the main categories cover all main aspects of the life of a prokaryotic organism: Cellular Processes, Metabolism, Information Processing, Lifestyles and Prophages and Mobile Genetic Elements. A last main category, Groups of Genes, assembles the genes/proteins based on the level of knowledge on the function, the localization and the nature of the gene product. This classification scheme is organism-independent and therefore, generally suitable for the systematic classification of protein function in bacteria. In line with this, the main category ‘Lifestyles’ covers species-specific functions and could be readily adapted to other organisms. With respect to the specificity of the subcategories, the classification scheme was designed to ensure the same level of detail at a certain sublevel; for example, all individual metabolic pathways (e.g. biosynthesis of amino acids or cell wall components) are classified as fourth level categories.
As mentioned above, the information about the cellular function (categories) and transcriptional regulation (regulons) is implemented in the SubtiWiki pages in two fields in the upper part of the gene pages: ‘Categories containing this gene/protein’ and ‘The gene is member of the following regulons’ (Figure 1). The respective categories and regulons are clickable, leading to the pages listing all category members at the third level of the classification and all regulon members, respectively (Figures 2 and 3). In the case of the functional categories, the corresponding main category and the directly higher-level category are displayed as ‘Parent categories’ and the other categories in the same branch of the tree as ‘Neighbouring Categories’ (Figure 3). Relevant publications are listed in the bottom part of the category and regulon pages, respectively. Overviews on all categories and regulons can be accessed from each SubtiWiki gene page.
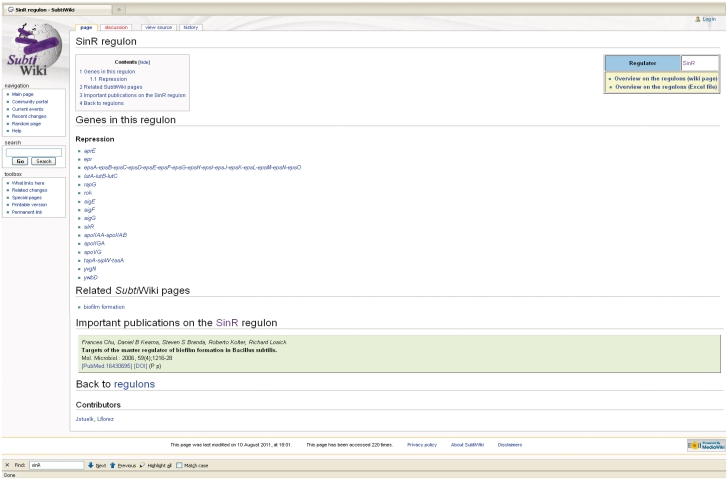
Figure 2.
Page on the SinR regulon in SubtiWiki. Regulon pages give an overview of all the genes that are regulated by a specific transcription factor. Additional links lead to related SubtiWiki pages and to publications connected to the chosen regulon. An overview of all regulons is available as SubtiWiki page or Excel file.
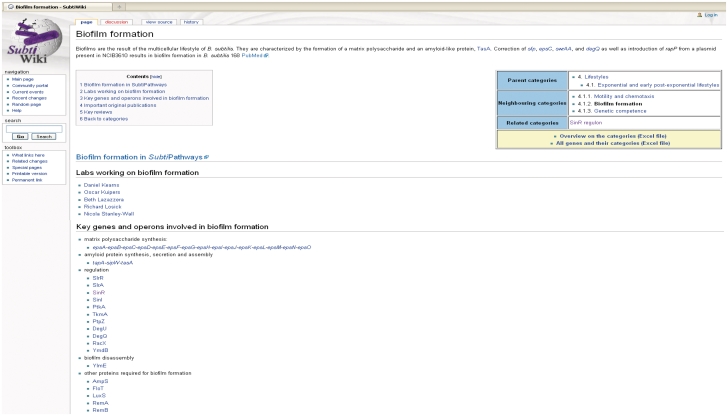
Figure 3.
Page for the category ‘Biofilm formation’ in SubtiWiki. All genes in the database are assigned to one or several functional categories. The main categories are divided into subcategories like Biofilm formation (part of the main category Lifestyles). The category pages list all the genes and operons that are involved in the given process, as well as links to key publications (not displayed).
Importantly, the functional and regulatory properties of a given gene are displayed together on the respective SubtiWiki gene page, thus providing an additional level of insight into the physiological role of a gene/protein of interest. For the first time, this compilation puts the claim of the ‘omics’ technologies, to provide information for discovering novel relationships into reality. As an example, the YxjG protein could be assigned as a putative methionine synthase based on the regulation that the yxjG gene shares with all other genes involved in methionine biosynthesis (20). Moreover, with the regulon gene lists and functional categories, it is now possible to directly access all genes/proteins that are related to each other by function, localization or regulation and to get a quick overview on any functional and regulatory aspect of B. subtilis.
As for the other content of SubtiWiki, the newly added types of information require regular updates, because the functional annotation of gene products, in particular those of so far unknown function, is continuously updated as more information is becoming available, which then also leads to the assignment to new or additional functional categories. The same holds true for the assignment of genes to transcription factor regulons.
In order to meet the specific requirements of bioinformatics applications, regularly updated versions of the following files are available for download: (i) the functional classification scheme, (ii) a table listing all categories with their respective genes and (iii) a table with all regulons and their member genes. The functional and regulatory classifications are extremely useful data sources for the analysis and interpretation of genome-scale experiments. First, they allow for data mining approaches such as functional profiling which use various kinds of prior knowledge for statistical enrichment analysis to infer physiological context of a list of genes or proteins. In addition, genome-scale experimental data can be visualized by displaying them on a functional classification or transcriptional regulatory annotation. For example, in a proteomic and transcriptomic profiling study of glucose-starved B. subtilis cells (21), Voronoi treemaps linking a representation of hierarchically structured functional categories or gene regulatory information (regulon/operon/gene) with gene expression data were used to support the analysis of a complementary proteome and transcriptome data set. Information about transcription factor regulons was derived from SubtiWiki. Moreover, analyses facilitating the understanding of the metabolic and regulatory network organization, such as the prediction of new transcription factor target genes and assignment of putative functions to so far un-annotated genes, are also supported by the SubtiWiki functional classification and our compilation of transcription factor regulons.
SubtiPathways and SubtInteract—two resources that complement SubtiWiki
Three genome-scale models of B. subtilis integrate the existing knowledge into models of metabolic and regulatory networks (22–24). Since these models are not easily accessible for the lab scientist, we developed a suite of diagrams of metabolism and regulation in B. subtilis, SubtiPathways. The SubtiPathways diagrams provide an interface for systems biology as they link information relevant for modelers and bench biologists. Today, SubtiPathways encompasses 35 diagrams that cover different aspects of B. subtilis physiology and regulation (Figure 4). These diagrams are linked to the SubtiWiki pages of the relevant genes and, on the other hand, each gene page contains a link to the specific diagram, if available.
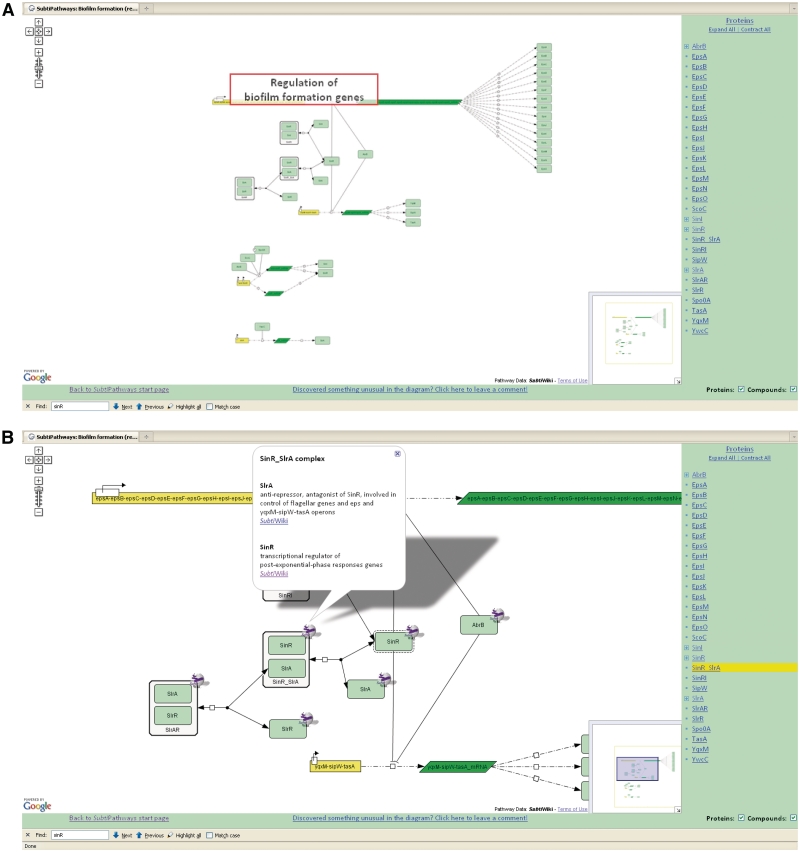
Figure 4.
Representation of Biofilm formation in SubtiPathways. Diagrams are available for a broad range of metabolic pathways and cellular processes (A). A list of all involved genes is shown on the right side and the diagram itself shows gene expression and regulation. At the highest magnification, a zoom function reveals clickable elements (B) that provide information on the protein (e.g. 3D structure, links to other pathways and SubtiWiki).
Recently, we have focused on protein–protein interactions. First, we generated a genome-scale model of the interactions using Cytoscape (25). This interaction model was then converted into a navigatable diagram using the Google maps API and the program CellPublisher (26). As described for SubtiPathways, the diagram can be intuitively navigated (by zooming and panning). Moreover, each protein in the diagram is directly linked to the corresponding SubtiWiki page (Figure 5A). In addition, we used our database that collects all interactions to generate protein-specific pages that display the interactions of this particular protein. In the left side bar, the complexity of the network can be selected, whereas links to the SubtiWiki gene pages are provided in the side bar on the right (Figure 5B). By September 2011, SubtInteract contained 1830 interactions involving 801 proteins and 5 RNAs. The protein-specific interaction networks are directly accessible from the Table on the top of the gene pages (Figure 1A).
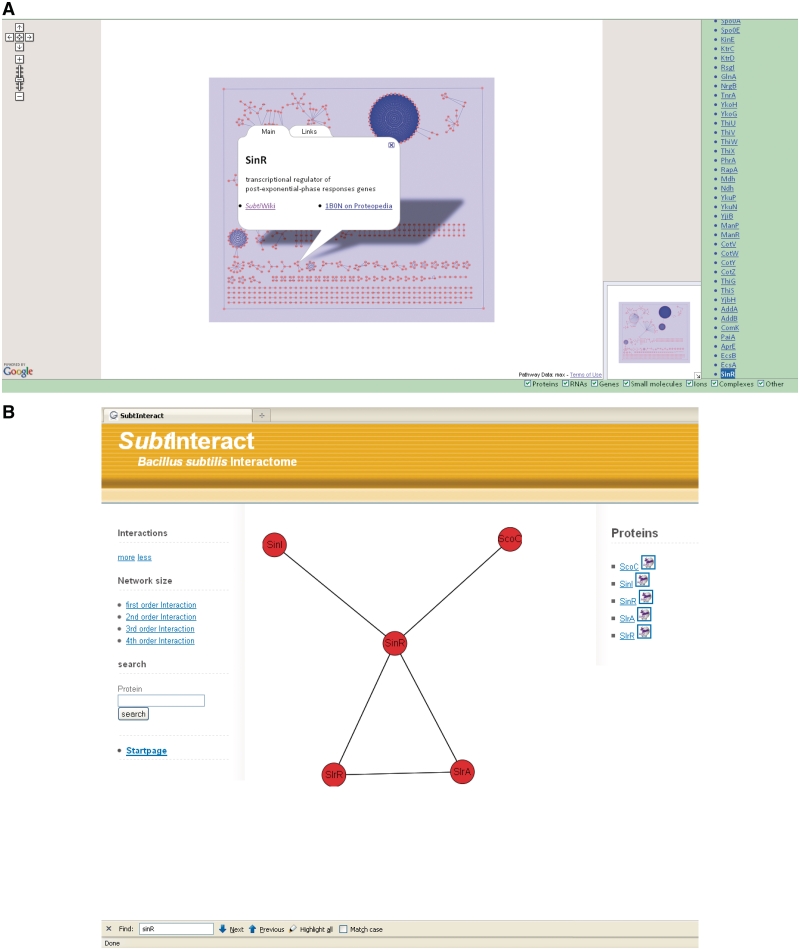
Figure 5.
Global and specific presentations in SubtInteract. The user interface allows a global view of protein interactions (A) with additional information on single proteins (e.g. SinR)-like protein structures and links to SubtiWiki or SubtiPathways. Furthermore, interaction partners of specified proteins can be shown with different network complexities (B).
PERSPECTIVES
SubtiWiki (with SubtiPathways and SubtInteract) has become one of the most complete inventories of knowledge on a living organism in one single resource. Both the continuous updates and the novel features contribute to its popularity that is reflected by more than one million page visits during the last 12 months. In the same period, 48 genes were given new designations in the scientific literature, and these new gene names were also adopted for SubtiWiki. Moreover, for 40 genes of previously unknown function, a functional annotation was assigned during the last year.
In the future, keeping up-to-date with the state of research will remain a key task for the development of SubtiWiki. In addition, we will link SubtiWiki to global gene expression data in order to provide a new type of information that is at the heart of the interest of many Bacillus researchers.
FUNDING
The Federal Ministry of Education and Research SYSMO network grant (PtJ-BIO/0315784B and 0315784A to J. S. and U.M.). Funding for open access charge: University of Göttingen
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We are grateful to Maximilian Fünfgeld for his help with the development of SubtInteract.
REFERENCES
- 1.Kunst F, Ogasawara N, Moszer I, Albertini AM, Alloni G, Azevedo V, Bertero MG, Bessières P, Bolotin A, Borchert S, et al. The complete genome sequence of the Gram-positive bacterium Bacillus subtilis. Nature. 1997;390:249–256. doi: 10.1038/36786. [DOI] [PubMed] [Google Scholar]
- 2.Moszer I, Glaser P, Danchin A. SubtiList: a relational database for the Bacillus subtilis genome. Microbiology. 1995;141:261–268. doi: 10.1099/13500872-141-2-261. [DOI] [PubMed] [Google Scholar]
- 3.Flórez LA, Roppel SF, Schmeisky AG, Lammers CR, Stülke J. A community-curated consensual annotation that is continuously updated: the Bacillus subtilis centred wiki SubtiWiki. Database. 2009 doi: 10.1093/database/bap012. doi: 10.1093/database/bap012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lammers CR, Flórez LA, Schmeisky AG, Roppel SF, Mäder U, Hamoen L, Stülke J. Connecting parts with processes: SubtiWiki and SubtiPathways integrate gene and pathway annotation for Bacillus subtilis. Microbiology. 2010;156:849–859. doi: 10.1099/mic.0.035790-0. [DOI] [PubMed] [Google Scholar]
- 5.Kitano H, Funahashi A, Matsuoka Y, Oda K. Using process diagrams for the graphical representation of biological networks. Nat. Biotechnol. 2005;23:961–966. doi: 10.1038/nbt1111. [DOI] [PubMed] [Google Scholar]
- 6.Marchadier E, Carballido-López R, Brinster S, Fabret C, Mervelet P, Bessières P, Noirot-Gros MF, Fromion V, Noirot P. An expanded protein-protein interaction network in Bacillus subtilis reveals a group of hubs: Exploration by an integrative approach. Proteomics. 2011;11:2981–2991. doi: 10.1002/pmic.201000791. [DOI] [PubMed] [Google Scholar]
- 7.Griffiths KK, Zhang J, Cowan AE, Yu J, Setlow P. Germination proteins in the inner membrane of dormant Bacillus subtilis spores colocalize in a discrete cluster. Mol. Microbiol. 2011;81:1061–1077. doi: 10.1111/j.1365-2958.2011.07753.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Delumeau O, Lecointe F, Muntel J, Guillot A, Guedon E, Monnet V, Hecker M, Becher D, Polard P, Noirot P. The dynamic partnership of RNA polymerase in Bacillus subtilis. Proteomics. 2011;11:2992–3001. doi: 10.1002/pmic.201000790. [DOI] [PubMed] [Google Scholar]
- 9.Sanders GM, Dallmann HG, McHenry CS. Reconstitution of the B. subtilis replisome with 13 proteins including two distinct replicases. Mol. Cell. 2010;37:273–281. doi: 10.1016/j.molcel.2009.12.025. [DOI] [PubMed] [Google Scholar]
- 10.Commichau FM, Rothe FM, Herzberg C, Wagner E, Hellwig D, Lehnik-Habrink M, Hammer E, Völker U, Stülke J. Novel activities of glycolytic enzymes in Bacillus subtilis. Mol. Cell. Proteomics. 2009;8:1350–1360. doi: 10.1074/mcp.M800546-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Meyer FM, Gerwig J, Hammer E, Herzberg C, Commichau FM, Völker U, Stülke J. Physical interactions between tricarboxylic acid cycle enzymes in Bacillus subtilis: evidence for a metabolon. Metab. Eng. 2011;13:18–27. doi: 10.1016/j.ymben.2010.10.001. [DOI] [PubMed] [Google Scholar]
- 12.Caspi R, Altman T, Dale JM, Dreher K, Fulcher CA, Gilham F, Kaipa P, Karthikeyan AS, Kothari A, Krummenacker M, et al. The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases. Nucleic Acids Res. 2010;38:D473–D479. doi: 10.1093/nar/gkp875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barbe V, Cruveiller S, Kunst F, Lenoble P, Meurice P, Sekowska A, Vallenet D, Wang T, Moszer I, Medigue C, et al. From a consortium sequence to a unified sequence: the Bacillus subtilis 168 reference genome a decade later. Microbiology. 2009;155:1758–1775. doi: 10.1099/mic.0.027839-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sierro N, Makita Y, de Hoon M, Nakai K. DBTBS: a database of transcriptional regulation in Bacillus subtilis containing upstream intergenic conservation information. Nucleic Acids Res. 2008;36:D93–D96. doi: 10.1093/nar/gkm910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moszer I, Kunst F, Danchin A. The European Bacillus subtilis genome sequencing project: current status and accessibility of the data from a new World Wide Web site. Microbiology. 1996;142:2987–2991. doi: 10.1099/13500872-142-11-2987. [DOI] [PubMed] [Google Scholar]
- 16.Riley M. Functions of the gene products of Escherichia coli. Microbiol. Rev. 1993;57:862–952. doi: 10.1128/mr.57.4.862-952.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ruepp A, Mewes HW. Prediction and classification of protein functions. Drug Discov. Today: Technol. 2006;3:145–151. doi: 10.1016/j.ddtec.2006.06.011. [DOI] [PubMed] [Google Scholar]
- 18.Ruepp A. The FunCat, a functional annotation scheme for systematic classification of proteins from whole genomes. Nucleic Acids Res. 2004;32:5539–5545. doi: 10.1093/nar/gkh894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Serres MH, Riley M. MultiFun, a multifunctional classification scheme for Escherichia coli K-12 gene products. Microb. Comparative Genomics. 2000;5:205–222. doi: 10.1089/omi.1.2000.5.205. [DOI] [PubMed] [Google Scholar]
- 20.Chi BK, Gronau K, Maeder U, Hessling B, Bacher D, Antelmann H. S-bacillithiolation protects against hypochlorite stress in Bacillus subtilis as revealed by transcriptomics and redox proteomics. Mol. Cell. Proteomics. 2011 doi: 10.1074/mcp.M111.009506. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Otto A, Bernhardt J, Meyer H, Schaffer M, Herbst F-A, Siebourg J, Mäder U, Lalk M, Hecker M, Becher D. Systems-wide temporal proteomic profiling in glucose-starved Bacillus subtilis. Nat. Commun. 2010;1:137. doi: 10.1038/ncomms1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oh YK, Palsson BO, Park SM, Schilling CH, Mahadevan R. Genome-scale reconstruction of metabolic network in Bacillus subtilis based on high-throughput phenotyping and gene essentiality data. J. Biol. Chem. 2007;282:28791–28799. doi: 10.1074/jbc.M703759200. [DOI] [PubMed] [Google Scholar]
- 23.Goelzer A, Bekkal Brikci F, Martin-Verstraete I, Noirot P, Bessières P, Aymerich S, Fromion V. Reconstruction and analysis of the genetic and metabolic regulatory networks of the central metabolism of Bacillus subtilis. BMC Syst. Biol. 2008;2:20. doi: 10.1186/1752-0509-2-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Henry CS, Zinner JF, Cohoon MP, Stevens RL. iBsu1103: a new genome-scale model of Bacillus subtilis based on SEED annotations. Genome Biol. 2009;10:R69. doi: 10.1186/gb-2009-10-6-r69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cline MS, Smoot M, Cerami E, Kuchinsky A, Landys N, Workman C, Christmas R, Avila-Campilo I, Creech M, Gross B, et al. Integration of biological networks and gene expression data using Cytoscape. Nat. Protoc. 2007;2:2366–2382. doi: 10.1038/nprot.2007.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Flórez LA, Lammers CR, Michna R, Stülke J. CellPublisher: a web platform for the intuitive visualization and sharing of metabolic, signaling and regulatory pathways. Bioinformatics. 2010;26:2997–2999. doi: 10.1093/bioinformatics/btq585. [DOI] [PubMed] [Google Scholar]