Abstract
FlyBase (http://flybase.org) is the leading database and web portal for genetic and genomic information on the fruit fly Drosophila melanogaster and related fly species. Whether you use the fruit fly as an experimental system or want to apply Drosophila biological knowledge to another field of study, FlyBase can help you successfully navigate the wealth of available Drosophila data. Here, we review the FlyBase web site with novice and less-experienced users of FlyBase in mind and point out recent developments stemming from the availability of genome-wide data from the modENCODE project. The first section of this paper explains the organization of the web site and describes the report pages available on FlyBase, focusing on the most popular, the Gene Report. The next section introduces some of the search tools available on FlyBase, in particular, our heavily used and recently redesigned search tool QuickSearch, found on the FlyBase homepage. The final section concerns genomic data, including recent modENCODE (http://www.modencode.org) data, available through our Genome Browser, GBrowse.
ORGANIZATION OF THE FLYBASE WEB SITE
The FlyBase web site is the portal for Drosophila-related information, currently containing 2.5 million pages covering 19 different data classes and 12 Drosophila reference sequenced genomes. Major sections include phenotypic, gene expression and interactions data curated from over 24 000 research papers as well as functional genomics projects. In addition, FlyBase maintains millions of links to other key resources such as strain and clone repositories, expression database (e.g. FlyExpress, http://www.flyexpress.net) and sequence databanks. The FlyBase homepage includes the QuickSearch query tool, which provides simple query access to the data in FlyBase, and clickable icons for easy access to other important search and display tools such as BLAST and GBrowse. The changing Commentary (http://flybase.org/static_pages/feature/previous/previous.html) highlights FlyBase news and improvements to the Drosophila community, while the left-hand sidebar carries fly-centric community news and conference information. The navigation bar hosts dropdown menus of query tools, files for download, taxonomic information on the genus Drosophila, documentation, Drosophila web resources, community news, help sections and links to previous releases of FlyBase. Of particular note is the ‘Documents’ menu, which contains release notes for each FlyBase release as well as our reference manual and guide to Drosophila nomenclature. In addition, the right-hand side of the navigation bar contains a small search box called ‘Jump to Gene’. This gene-specific search was designed for fast navigation to a gene report, and works optimally when the precise gene symbol is entered. Finally, there is a ‘Contact FlyBase’ link in the footer of every FlyBase web page for users to submit questions or comments about FlyBase.
FlyBase reports
Data in FlyBase are organized into report types corresponding to different data classes. There are 19 FlyBase data classes for which reports are currently available, listed in Table 1, along with URLs for example reports. FlyBase reports are web pages that bring together all of the information for one object of a given data class. For example, there are reports for genes (e.g. hedgehog—hh), alleles (e.g. cnnKG05783) and cell lines (e.g. Schneider's-line-2). Each FlyBase report begins with a section of basic information applicable to most records of the class and ends with a list of references from which the report is composed. Most of this top-level information is standardized and makes use of Controlled Vocabularies (CVs) enabling automated retrieval of the data (see later for further information on the use of controlled vocabularies in searching). Further content is found within nested subsections that can be displayed or hidden according to your interest. Recent updates or changes to a report are highlighted in green. Reports incorporate links to related FlyBase reports and graphical displays, and link-outs to external sources of information. A help page (http://flybase.org/static_pages/newhelp/report_help.html) for each report class, found via the Help button at the top of the report, describes the contents of each report field. Becoming familiar with report fields will aid browsing for particular types of data and using our advanced field-aware search tools such as QueryBuilder, which target queries to specific types of information.
Table 1.
Reports in FlyBase
| FlyBase report | ID Prefix | No. of reports | Example |
|---|---|---|---|
| Aberration | FBab | 22 095 | Df(3R)BSC678 |
| Allele | Fbal | 174,584 | dppEP2232 |
| Balancer | FBba | 562 | TM6B |
| Cell line | FBtc | 140 | S2R+ |
| Clone | FBcl | 722 146 | GH07782 |
| Gene | FBgn | 218 959 | dpp |
| Image | FBim | 981 | Hartenstein (1993) Tracheal System Adult |
| Insertion | FBti | 143 176 | P{EP}dppEP2232 |
| Interactions | FBig | 59 755 | Physical association between Prosβ3 and Pros29 |
| Library collection | FBlc | 416 | TRiP-4 |
| Natural transposon | FBte | 1391 | P-element |
| Polypeptide | FBpp | 203 595 | dpp-PB |
| Recombinant construct | FBmc/FBms | 3072 | pGaTN |
| References | FBrf | 198 436 | St. Pierre and McQuilton, P. (2009) |
| Sequence feature | FBsf | 383 313 | ras-protein_bind-1 |
| Stock | FBst | 113 292 | w[1118]; P{GD10831}v26115 |
| Term report | GO/FBbt/FBcv/FBdv/SO | 335 609 | increased cell number |
| Transcript | FBtr | 215 074 | dpp-RB |
| Transposon | FBtp | 67 576 | P{UAS-dpp.GFP.T} |
An example: the gene report
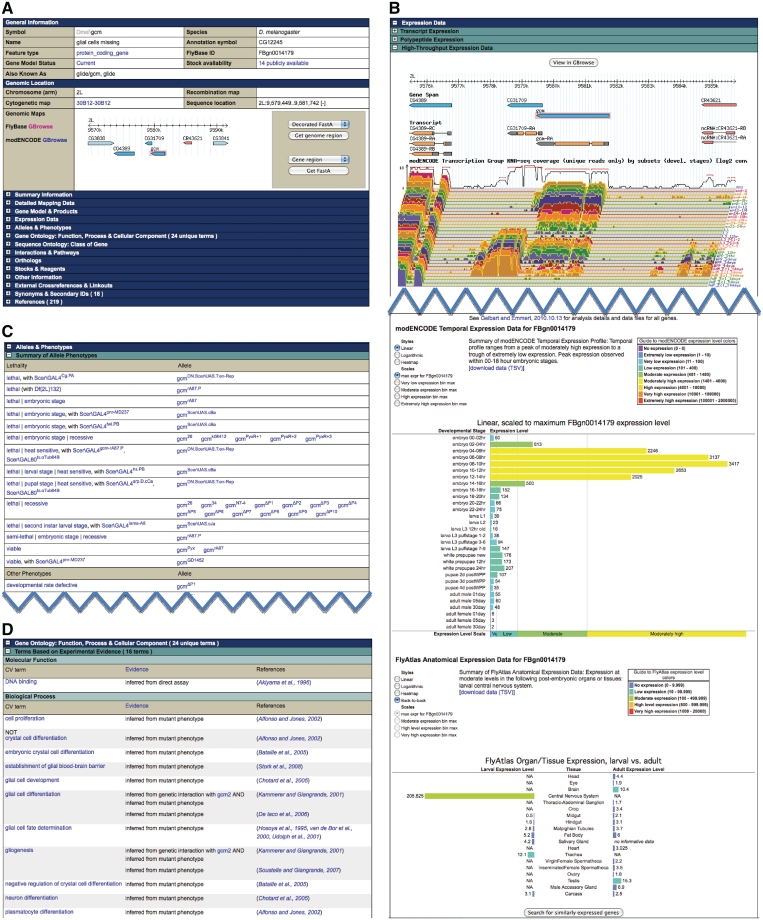
Much core information within FlyBase is found in, or linked to, the Gene Reports. The glial cells missing (gcm) Gene Report (http://flybase.org/reports/FBgn0014179.html) is illustrated in Figure 1. The report is split into 17 sections, with further subsections that can be clicked open to reveal specific data. The ‘General Information and Genomic Locations’ sections at the top of the report contain the gene symbol, name, common synonyms, a small ‘GBrowse’ image and provide access to the sequence and a list of related stocks, when available. Beneath this, there is a ‘Summary Information’ section with a computed prose summary of the gene report, providing information on gene function, protein domains, numbers of alleles and transcripts and further summaries of the computed and curated information available about the gene and its products. Curated prose gene summaries created by external projects such as Interactive Fly (http://www.sdbonline.org/fly/aimain/1aahome.htm) are also included in this section when available. These summaries are followed by a list of recent updates (i.e. those made in the last few releases), the data-containing sections and, finally, as with all reports, a list of the associated references.
Figure 1.
Example Gene Report. (A) The General Information and Genomic Location sections, found at the top of all reports. (B) High-Throughput Expression data section. (C) GO data report section. (D) Alleles & Phenotype section, with links to both the phenotype controlled vocabulary term report (where you can find a definition and search for other alleles annotated with this term) and each allele report.
Example Gene Report sections
Gene expression and phenotype data are found in the ‘Expression Data’ and ‘Alleles & Phenotypes’ sections of the Gene Report (highlighted in Figure 1). The Expression Data section contains four subsections. The first two (‘Transcript Expression’ and ‘Polypeptide Expression’) contain data extracted manually by FlyBase curators from publications and personal communications. Controlled Vocabulary terms describe the stage and tissue/position where expression is found, along with the reference in which these data are reported. Third is the new ‘High-Throughput Expression Data’ subsection (Figure 1B) that houses a wealth of temporal and anatomical data. These data are displayed through various graphs to allow an instant visual understanding of the data. A handy way to search the expression data from the gene report page is the ‘Search for similarly expressed genes’ button, found at the bottom of the ‘High-Throughput Expression Data’ section. The final subsection, ‘External Data and Images’, provides link-outs (when available) to external web sites, such as FlyExpress.
The ‘Alleles & Phenotypes’ section (Figure 1C) is split into five subsections. The ‘Summary of Allele Phenotypes’ subsection details curated CV terms alongside the alleles with which they are associated. Controlled Vocabularies allow curators to describe the various features and processes that biologists study in a consistent way, which helps to clarify the data and supports effective searching. By using CVs, it becomes possible to search for genes and other specific data classes associated with various equivalent descriptions such as ‘translation’ and ‘protein synthesis’, or ‘wing disc’ and ‘mesothoracic disc’. This section is useful when you wish to see an overview of the phenotypes associated with alleles of a gene. Clicking on any CV term or allele takes you to the appropriate term or allele report. Below this summary section, more detailed phenotype descriptions are split between classical alleles and those alleles carried on transgenic constructs. As with the summary phenotypes, these data are presented in a table with clickable links to the allele and CV term reports if you wish to explore further. Aneuploid aberrations, transgenic constructs and insertions are also referenced in this section.
Gene Ontology [GO (1), http://www.geneontology.org/) annotations are also provided on the Gene Report page (Figure 1D) and are divided into two subsections. The first shows those terms from the three main categories of Gene Ontology (Molecular Function, Cellular Component and Biological Process) assigned from experimental analysis of the gene/gene product. The second shows those terms based on predictions or assertions (such as via sequence similarity to a gene of known function).
The Gene Report is perhaps the most important report in FlyBase, as it acts as a portal for further exploration of the data within FlyBase. The Gene Report provides many links into other FlyBase reports, as well as to external databases. This includes links to sequence reports for the gene in multiple nucleotide and protein sequence databases (e.g. RefSeq, GenBank and UniProt), link-outs to other content providers (e.g. Interactive Fly and FlyExpress) and links to other databases containing functional data (e.g. BioGRID and FlyMine). A list of all the link-outs, plus short descriptions of each field in the gene report, is provided in the gene report help page (http://flybase.org/static_pages/newhelp/gene_help.html), found in the report help section in the help menu.
SEARCHING FLYBASE
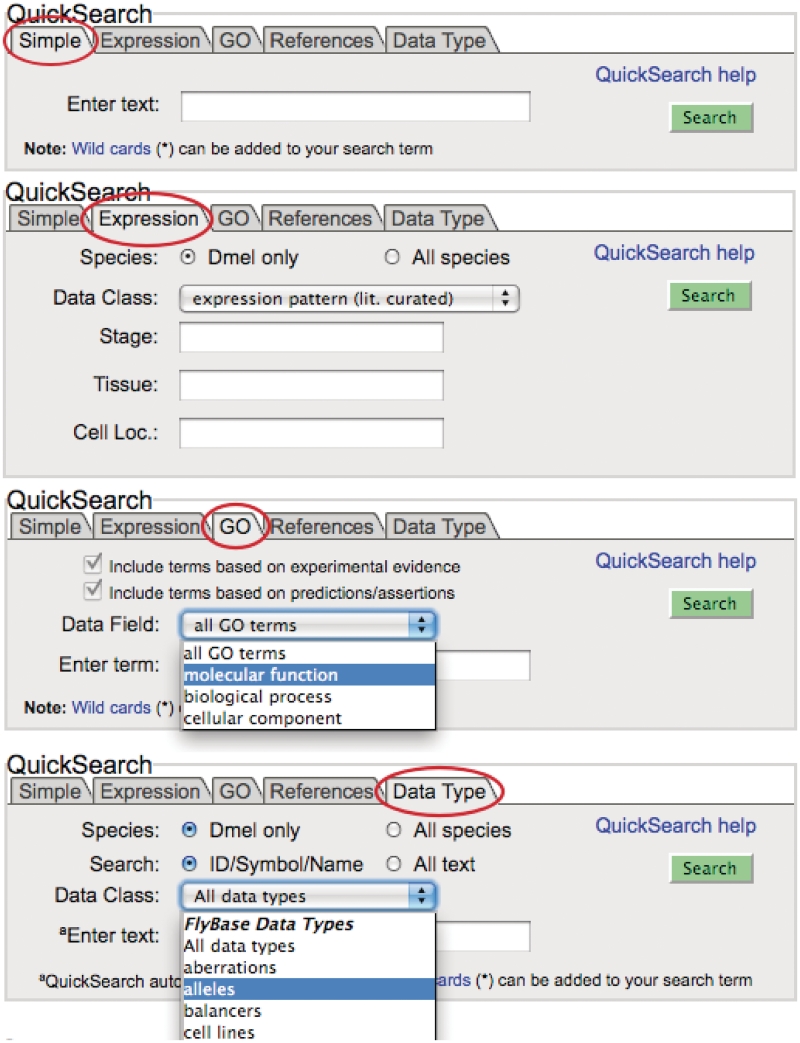
How do you find reports containing the information that you want? Most often, the search tool QuickSearch will be the best place to start a search (Figure 2). This section describes the updated version of ‘QuickSearch’ while the following section, ‘Alternatives to QuickSearch’, details some of the other tools available on FlyBase.
Figure 2.
QuickSearch.
QuickSearch
The QuickSearch tool on the FlyBase home page has recently been updated with extended capabilities. Forms for searching specific types of data have been separated into ‘tabs’, arrayed at the top of the QuickSearch window. This tabbed organization has allowed us to make each search form clearer, and in most tabs we have been able to add extra functionality. Several of the tabs contain entirely new search tools, such as a new ‘Simple’ search form, an easy-to-use tool with access to all the data types in FlyBase (see Figure 2).
The ‘Simple’ tab performs a global search of FlyBase data (Figure 2 top). The form has a very clean interface, with only a textbox and a ‘Search’ button for input. When a single word or phrase is entered, this search combs all the FlyBase data records that can be text-searched and returns a result page summarizing the matching records by data type. Clicking on one of these data types takes you to a table of individual matches in that data type. Alternatively, you can edit the phrase directly and search again, without having to start over.
There are several tabs on QuickSearch that allow searches using controlled vocabulary terms. These tabs provide intuitive domain-specific searches of FlyBase reports based on GO terms, anatomical, developmental-stage-specific or phenotypic class terms used to annotate phenotypes and anatomical and/or developmental-stage-specific terms used to annotate gene expression. Combinations of CV terms can be searched using the forms in these tabs
The ‘References’ tab offers a search of the extensive FlyBase bibliography. Searches can be filtered by title/abstract text, journal name, publication type and reference IDs (PubMed or FlyBase), in addition to the author and date filters. Appropriate fields also allow the use of Boolean operators, so you can search for papers authored by e.g. ‘Smith NOT Johnson’ or published ‘>2006’ (after 2006).
The ‘Data Type’ tab contains a trimmed-down version of the previous QuickSearch form. The lengthy drop-down menu of data classes has been shortened considerably, with many of the data classes now having their own tabs, but the behavior of the search in this tab is otherwise largely unchanged. Here you can search specific data classes in the FlyBase database, such as stocks, gene associations, sequence features or aberrations. When you search any one of the FlyBase data classes, your results will be restricted to only those hits from within that data class. If you are unsure how FlyBase classifies the item you are looking for (e.g., a gene, allele, insertion or clone), you can select the ‘All data types’ option to have QuickSearch search every class of data in FlyBase (or use the ‘Simple’ tab to search all report data in FlyBase).
Many of the tabs make use of our FlyBase-specific auto-complete feature. Auto-completion is probably familiar to Google™ (or similar) web search page users, and most browsers now have a mechanism like this to provide hints when users are filling out forms. The textboxes in auto-complete-enabled QuickSearch tabs suggest search phrases that are specific to FlyBase data reports. This auto-complete feature overrides your browser's auto-complete function.
An advanced coordinated auto-complete has been active in the QuickSearch tool for some time. Here is an example of how it works in the ‘Expression’ tab:
When the ‘expression pattern (lit. curated)’ data class is selected, text box fields for Stage, Tissue and Cell Loc. (cell location) are displayed. The auto-complete for these three fields is coordinated in the following sense: Suppose you enter ‘fertilized egg stage’ in the Stage text box. When you move your focus to the Tissue text box, auto-complete there will show only four options; ‘egg’, ‘female pronucleus’, ‘fertilized egg’ and ‘male pronucleus’. This is because, out of the multitude of CV terms available for the Tissue field, only these four terms have actually been used in combination with ‘fertilized egg stage’ by curators in an annotation captured in the FlyBase database. If you enter any other term in the Tissue text box, even though it may be a valid CV term for that field, your search would return zero hits, because there are no FlyBase reports containing that combination of CV terms.
Using the terms suggested by the auto-complete feature ensures that you do not enter terms that would be mutually exclusive (or have simply not been used by curators). Terms suggested by the auto-complete should always return results. If the coordinated auto-complete does not offer a term you wish to enter in a field, it is because this term does not appear in combination with some other term you have entered elsewhere on the form. In this case, you should try another combination.
Many will find that the search capabilities of QuickSearch meet your needs and we recommend the tool as the first entry point to FlyBase data. More complex tools have a steeper learning curve, but ultimately allow for very powerful searches across all of FlyBase. To become familiar with the tools’ capabilities and to learn how best to use them may require some investment of time, but these tools allow very efficient filtering of the vast amounts of Drosophila data. A list of all the tools, with descriptions of the data they search, can be found in the tools overview section (http://flybase.org/static_pages/docs/tools_overview.html), found under the tools menu in the FlyBase navigation bar.
Alternatives to QuickSearch
While QuickSearch offers rapid, intuitive methods to search FlyBase, some users will want to delve more deeply or obtain a more focused result set than QuickSearch can achieve. For those users FlyBase provides more robust search tools, such as BLAST, QueryBuilder, or TermLink.
The FlyBase ‘BLAST’ tool (http://flybase.org/blast/) is an ideal entry point for researchers interested in the fly homolog of their favorite non-Drosophila gene. It will retrieve Drosophila genes with sequences similar to the submitted sequence.
‘QueryBuilder’ (http://flybase.org/.bin/qbgui.fr.html) is a web-based tool to build powerful queries across the many data types in FlyBase. With experience, one can construct queries to obtain hit lists of genes and other data types matching almost any set of criteria. In addition, there are a number of pre-defined QueryBuilder templates available to guide the less-experienced user through the use of combinatorial searches between and within report pages.
The ‘TermLink’ (http://flybase.org/static_pages/termlink/termlink.html) tool enables browsing of the CVs used by FlyBase curators and the retrieval of CV term reports. These reports contain a definition for the term (when available), a relationship tree showing similar terms, as well as links to curated data (phenotypes, expression patterns etc.) described using the term.
Batch download
You may wish to export or download a list of genes or other items generated using one of the search methods described above. FlyBase offers several means to do this.
When using QuickSearch, a result ‘hit list’ page will have two buttons at the top: a ‘Results Analysis/Refinement’ button and a ‘Hitlist Conversion Tools’ button. The first button displays an assortment of ways to summarize and analyze the result list. The second displays a similar variety of ways to export or download the list. For example, one can download a text document containing the FlyBase ID numbers (e.g. FBgn0261526) for every item in your list, or export a hit list to the Batch Download tool.
The ‘Batch Download’ tool (http://flybase.org/static_pages/downloads/ID.html) allows you to save a file containing information about each item in the list. The tool provides options that depend on the type of data in the list. For instance, if you have obtained a list of gene reports, the Batch Download tool will give you the option to download a FASTA file with each gene sequence. Likewise, if you have a list of references from the FlyBase bibliography, the Batch Download tool will let you download the references in one of several tabular formats.
The FlyBase site also provides many precomputed bulk data files for simple download. Data files that have been requested frequently by FlyBase users are compiled at each release, using the current release data, and links to these have been collected in the Precomputed Files page. Using the navigation bar, go to Files -> Files Overview for a description of the available files or to Files -> Precomputed files (http://flybase.org/static_pages/downloads/bulkdata7.html) for direct access.
GENOMES IN FLYBASE: USING GBROWSE
GBrowse (2) is a GMOD (Generic Model Organism Database, http://gmod.org) tool that displays customizable genomic features along the chromosomal axis. In the customized FlyBase version of GBrowse, the primary genome is that of Drosophila melanogaster. However, the genomes of several other insects and model organisms are available for browsing as well.
Features and tracks
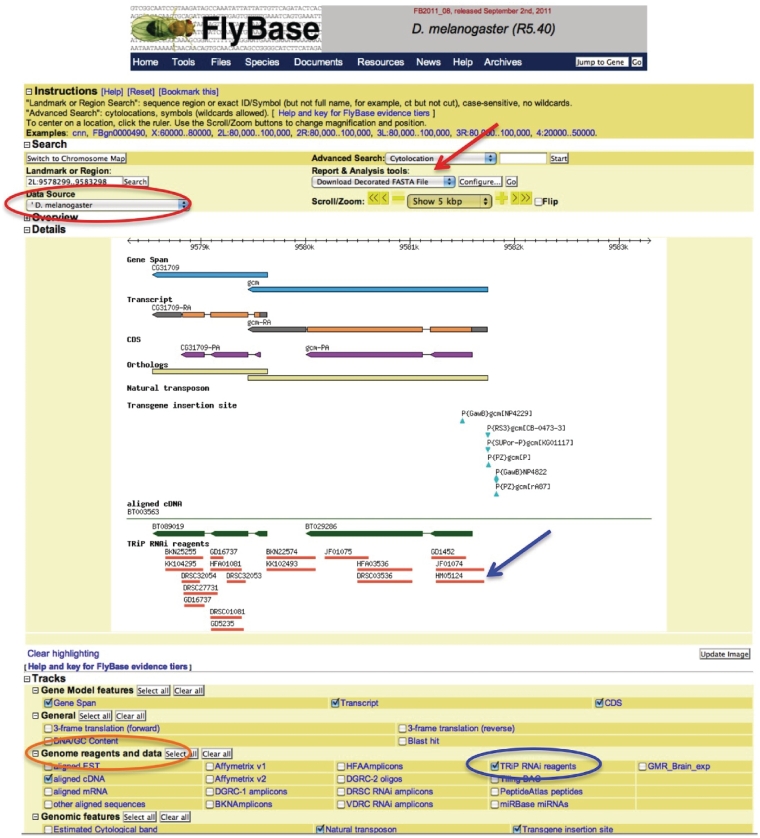
Genome features are bits of sequence that have been either assigned a function (e.g. transcription factor binding sites and exon junctions) or reported to be the location of an event (e.g. a mutation or transgenic insertion). A group of features of a certain type is called a ‘track’. For example, all exon junctions for all transcripts are grouped into the same track, named ‘RNA-Seq exon junctions’. Some tracks are visible by default, but to see additional tracks, you will need to click the appropriate checkbox and update the GBrowse image. The tracks are grouped and listed by title beneath the GBrowse image (Figure 3).
Figure 3.
GBrowse. The Data Source menu (red circle, top left), the Track Groupings (orange circle, bottom left) and the Feature Tracks (blue circle, bottom right) are shown. The features (members of the feature track) are indicated with a blue arrow. The location of the ‘table view’ option is indicated with a red arrow.
Navigating GBrowse
The Data Source menu (Figure 3) contains a drop-down list of all the GBrowse data sources. The top items are all based upon the D. melanogaster genome. Feature tracks are divided into different GBrowse views to efficiently utilize the space and optimize performance. The default view contains feature tracks related to gene model annotation (e.g. aligned cDNAs and gene predictions). One can switch to a different genome view by selection from the Data Source menu. Other D. melanogaster views include RNA-Seq expression data, Stocks and Reagents and various large-scale modENCODE data sets (Table 2). The Data Source menu also contains the genomes for other sequenced drosophilids and other model organisms.
Table 2.
modENCODE (and related) data sets integrated into FlyBase (as of FB2011_08)
| Data set | Contributors | Library/Collection report |
|---|---|---|
| RNA-Seq based exon junctions | Graveley et al., 2011 (modENCODE) | FBlc0000203 (http://flybase.org/reports/FBlc0000203.html) |
| Daines et al., 2011 (Baylor College of Medicine) | FBlc0000204 (http://flybase.org/reports/FBlc0000204.html) | |
| Insulators class I | Negre et al., 2011 | FBlc0000058 (http://flybase.org/reports/FBlc0000058.html) |
| Insulators class II | Negre et al., 2011 | FBlc0000059 (http://flybase.org/reports/FBlc0000059.html) |
| Chromatin domains | modENCODE Consortium, Roy, S., et al., 2010 | FBlc0000199 (http://flybase.org/reports/FBlc0000199.html) FBlc0000200 (http://flybase.org/reports/FBlc0000200.html) |
| Kharchenko et al., 2011 | FBlc0000199 (http://flybase.org/reports/FBlc0000199.html) FBlc0000200 (http://flybase.org/reports/FBlc0000200.html) | |
| Filion et al., 2010 | FBlc0000187 (http://flybase.org/reports/FBlc0000187.html) | |
| RNA-Seq expression data (developmental profile) | Graveley et al., 2011 | FBlc0000085 (http://flybase.org/reports/FBlc0000085.html) |
| Daines, et al., 2011 | FBlc0000060 (http://flybase.org/reports/FBlc0000060.html) | |
| RNA-Seq expression data (treatments) | Graveley et al., 2011 | FBlc0000236 (http://flybase.org/reports/FBlc0000236.html) |
| RNA-Seq expression data (cell lines) | Graveley et al., 2011 | FBlc0000260 (http://flybase.org/reports/FBlc0000260.html) |
| RNA-Seq expression data (tissues) | Graveley et al., 2011 | FBlc0000206 (http://flybase.org/reports/FBlc0000206.html) |
| RNA A-I editing sites | Graveley et al., 2011 | FBlc0000259 (http://flybase.org/reports/FBlc0000259.html) |
| Transcription factor binding sites | modENCODE Consortium, Roy, S., et al., 2010 | FBlc0000258 (http://flybase.org/reports/FBlc0000258.html) |
| Origins of Replication (Orc2 binding sites) | Eaton et al., 2011 | FBlc0000189 (http://flybase.org/reports/FBlc0000189.html) FBlc0000188 (http://flybase.org/reports/FBlc0000188.html) |
| Embryonic Enhancers | Negre et al., 2011 | FBlc0000415 (http://flybase.org/reports/FBlc0000415.html) |
| Embryonic PRE Silencers | Negre et al., 2011 | FBlc0000414 (http://flybase.org/reports/FBlc0000414.html) |
To navigate to a particular region of the genome, gene symbols (e.g. ade2 and CG31643), feature symbols or IDs (e.g. P{EP}dppEP2232 and FBti0010414) or sequence ranges (e.g. 2L:2428454..2459609) can be entered in the Landmark or Region search box (at the upper left of the window). The Search box is auto-complete enabled, but the search feature on GBrowse is not as sophisticated as that for our other tools, such as QuickSearch. If you are unsure of the symbol you are searching for, we recommend you start by using QuickSearch to identify your gene or feature of interest before attempting to find it in GBrowse.
GBrowse data can also be viewed in a table format. Using this view allows you to see the sequence coordinates for all the features within a region and provides a compact way to display and/or print the data contained within a particular GBrowse view. The table view option can be selected from the ‘Report & Analysis tools’ drop-down menu in the ‘Search’ section.
Integration with FlyBase reports
While GBrowse provides a visual representation of data, the descriptions, relationships and attributions of the data reside on the related report pages. For example, in GBrowse you can see the insertion site of a P-element transgene and can immediately determine its relationship to nearby transcripts. To find out what is known about that insertion you can view the associated report. Most GBrowse features are linked directly to their reports through a single click. Reciprocally, those report pages concerning features that can be mapped to the genome have links (via the small GBrowse glyphs) directly to the relevant genomic location in GBrowse. In addition, for several types of features, including insertions, mousing-over the glyph in GBrowse produces a pop-up containing useful information.
Some data sets are displayed only on GBrowse without any companion report pages. Examples of such data sets include RNA-Seq coverage data and chromatin domains. For these data, explanations of the experiments performed can be found on the associated library/collection reports that are linked to the track name (Table 2) (3–8).
modENCODE data
Data from the modENCODE projects (9) can be found on GBrowse in the relevant data source views. In addition to the extensive collection of gene expression data, the modENCODE project has also produced several sets of data relevant to how genes interact (e.g. transcription factor binding sites) and are regulated (e.g. insulators, RNAi editing sites). Descriptions of these data sets can be found on the relevant library/collection report (shown in Table 2); descriptions of the individual sequence features can be found on the relevant sequence feature reports (linked to the feature glyph in GBrowse).
Through GBrowse, we can begin to grasp the complexity of the genome. Visual representation of data makes GBrowse an excellent starting point for studying a region of interest, and GBrowse can also serve as a visual summary of data related to a gene of interest. Direct links between GBrowse and FlyBase reports allow GBrowse to remain as compact and intuitive as possible while ensuring that the wealth of descriptive data available for the drosophilid genomes can be easily accessed.
FLYBASE AND THE FLY COMMUNITY
We suggest FlyBase be referenced in publications by citing this publication and the FlyBase URL (http://flybase.org). We also recommend that when you are using FlyBase data (in your notebooks, spreadsheets, papers etc.) you make note of the FlyBase web site release (e.g. FB2011_08; the current release can be found in the header and footer on every page) and/or the sequenced species assembly.version release (e.g. D. melanogaster R5.40, found in the GBrowse header). In addition, we recommend that authors incorporate FlyBase object identifiers (e.g. FBgn and FBal) in addition to symbols for the unambiguous identification of intended FlyBase entities. Finally, we suggest that when preparing supplementary materials, you provide tabular data either in tab-separated files or in a spreadsheet rather than a PDF. Following these recommendations will greatly aid FlyBase curators in integrating your data into FlyBase.
FUNDING
National Human Genome Research Institute at the National Institutes of Health (P41 HG00739); and Medical Research Council (UK) (G1000968). Funding for open access charges: NIH NHGRI grant.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We would like to thank the PIs, curators and developers of FlyBase for their comments on the manuscript. The Current FlyBase Consortium comprises: William Gelbart, Nick Brown, Thomas Kaufman, Kathy Matthews, Maggie Werner-Washburne, Richard Cripps, Lynn Crosby, Adam Dirkmaat, David Emmert, L. Sian Gramates, Kathleen Falls, Beverley Matthews, Susan Russo, Andy Schroeder, Susan St Pierre, Pinglei Zhou, Mark Zytkovicz, Boris Adryan, Stephanie Bunt, Marta Costa, Helen Field, Steven Marygold, Peter McQuilton, Gillian Millburn, Laura Ponting, David Osumi-Sutherland, Ray Stefancsik, Susan Tweedie, Helen Atrill, Josh Goodman, Gary Grumbling, Victor Strelets, Jim Thurmond, J.D. Wong, Harriett Platero.
REFERENCES
- 1.The Gene Ontology Consortium. Gene ontology: tool for the unification of biology. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stein LD, Mungall C, Shu S, Caudy M, Mangone M, Day A, Nickerson E, Stajich JE, Harris TW, Arva A, et al. The generic genome browser: a building block for a model organism system database. Genome Res. 2002;12:1599–1610. doi: 10.1101/gr.403602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Graveley BR, Brooks AN, Carlson JW, Duff MO, Landolin JM, Yang L, Artieri CG, van Baren MJ, Boley N, Booth BW, et al. The developmental transcriptome of Drosophila melanogaster. Nature. 2011;471:473–479. doi: 10.1038/nature09715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Daines B, Wang H, Wang L, Li Y, Han Y, Emmert D, Gelbart W, Wang X, Li W, Gibbs R, et al. The Drosophila melanogaster transcriptome by paired-end RNA sequencing. Genome Res. 2011;21:315–324. doi: 10.1101/gr.107854.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nègre N, Brown CD, Ma L, Bristow CA, Miller SW, Wagner U, Kheradpour P, Eaton ML, Loriaux P, Sealfon R, et al. A cis-regulatory map of the Drosophila genome. Nature. 2011;471:527–531. doi: 10.1038/nature09990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kharchenko PV, Alekseyenko AA, Schwartz YB, Minoda A, Riddle NC, Ernst J, Sabo PJ, Larschan E, Gorchakov AA, Gu T, et al. Comprehensive analysis of the chromatin landscape in Drosophila melanogaster. Nature. 2011;471:480–485. doi: 10.1038/nature09725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Filion GJ, van Bemmel JG, Braunschweig U, Talhout W, Kind J, Ward LD, Brugman W, de Castro IJ, Kerkhoven RM, Bussemaker HJ, et al. Systematic protein location mapping reveals five principal chromatin types in Drosophila cells. Cell. 2010;143:212–224. doi: 10.1016/j.cell.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eaton ML, Prinz JA, Macalpine HK, Tretyakov G, Kharchenko PV, Macalpine DM. Chromatin signatures of the Drosophila replication program. Genome Res. 2011;21:164–174. doi: 10.1101/gr.116038.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.modENCODE Consortium—Roy S, Ernst J, Kharchenko PV, Kheradpour P, Nègre N, Eaton ML, Landolin JM, Bristow CA, Ma L, Lin MF, et al. Identification of functional elements and regulatory circuits by Drosophila modENCODE. Science. 2010;330:1787–1797. doi: 10.1126/science.1198374. [DOI] [PMC free article] [PubMed] [Google Scholar]