Abstract
zfishbook is an internet-based openly accessible database of revertible protein trap gene-breaking transposon (GBT) insertional mutants in the zebrafish, Danio rerio. In these lines, a monomeric red fluorescent protein (mRFP) is encoded by an artificial 3′ exon, resulting in a translational fusion to endogenous loci. The natural transparency of the zebrafish embryo and larvae greatly facilitates the expression annotation of tagged loci using new capillary-based SCORE imaging methods. Molecular annotation of each line is facilitated by cloning methods such as 5′-Rapid Amplification of cDNA Ends (RACE) and inverse polymerase chain reaction (PCR). zfishbook (http://zfishbook.org) represents a central hub for molecular, expression and mutational information about GBT lines from the International Zebrafish Protein Trap Consortium (IZPTC) that includes researchers from around the globe. zfishbook is open to community-wide contributions including expression and functional annotation. zfishbook also represents a central location for information on how to obtain these lines from diverse members of the IZPTC and integration within other zebrafish community databases including Zebrafish Information Network (ZFIN), Ensembl and National Center for Biotechnology Information.
INTRODUCTION
Understanding gene function in vivo is the next step in the post-genome era. Zebrafish (Danio rerio) is an excellent model of vertebrate developmental biology. Continued application of zebrafish to study the genetics and biology of cancer, behavior and development has greatly increased the overall utility of this model of vertebrate biology. Transposons are powerful tools to integrate DNA into zebrafish genes (1–3). Gene-breaking transposons (GBTs) are effective insertional mutagens in the zebrafish (4–6). These mutagenic transposons now include a protein trap that creates a fusion between the interrupted gene product and a monomeric red fluorescent protein (mRFP) while simultaneously depleting the expression of the native gene transcript to levels generally <1% of normal (6). In summary, these transposon insertions rapidly tag transcription units (genes) for easy cloning, provide dynamic fluorescence expression representing gene expression domains or protein trafficking, while producing mutations in genes that permit functional annotation of the disrupted gene. Each GBT mutation is also conditionally reversible using Cre recombinase or transient delivery of a splice-blocking morpholino.
SIGNIFICANCE
The International Zebrafish Protein Trap Consortium (IZPTC) was launched with over 325 active or cryopreserved lines at the Mayo Clinic with funding to produce another 1000 lines over the next 3 years. Currently, this international team includes members from Canada, Germany, India, Singapore and the USA with the shared goal to dramatically increase access to new GBT zebrafish lines for the research community. These GBT mutants represent the first collection of engineered conditional mutants outside of the mouse system. Zebrafish are an excellent system for recording real-time, dynamic expression of mRFP-tagged fusion proteins because of its external development and optical clarity, providing novel expression data directly associated with function in a living vertebrate.
REPRESENTATIVE DATA
Overview
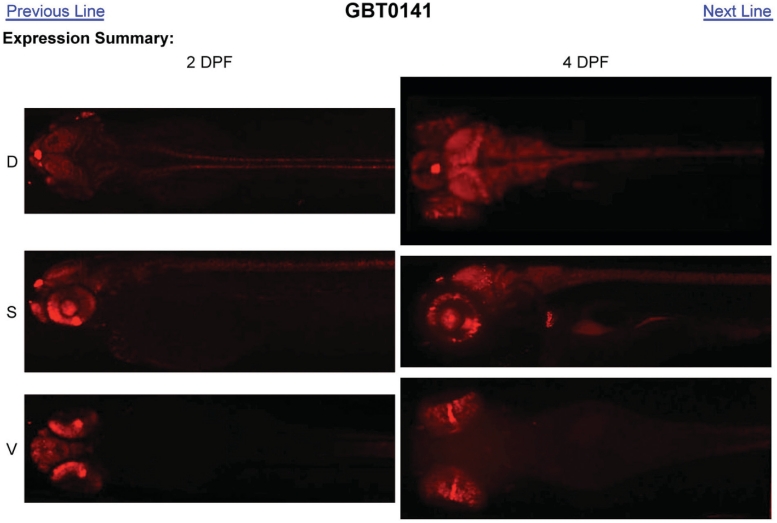
Each GBT line has its own digital notebook. The header for the notebook summarizes known information for the line including allele information, expression and phenotype data, and molecular data including sequence tags and primer sequences (Figure 1). The allele information includes the official GBT line designation, inventory status, source laboratory, mutagenesis vector used, tagged gene name and status, alternate names from source laboratory and the official Zebrafish Information Network (ZFIN) allele designation. For expression and phenotype data, a link to the full media library, a list of Zebrafish Anatomical (ZFA) Ontology expression tags annotated in the mRFP fusion protein images, and information about observed phenotypes or homozygous mutant viability is available within the summary block. Just below the summary block there are links to previous and next lines, followed by an expression summary at 2 and 4 days post-fertilization (DPF) (Figure 2).
Figure 1.
Example GBT summary block—GBT0141. At the top of each GBT line digital notebook is a summary block. The summary block contains the following elements. (A) Specific GBT line information including the official GBT line designation, inventory status, source laboratory, mutagenesis vector used, tagged gene name and status, alternate names from the source laboratory and the official ZFIN allele designation. (B) Expression and phenotype data including a link to the line's media gallery, a list of annotated ZFA Ontology tags, and a description of any known phenotype(s). (C) Links to molecular data including sequence tags, external database links and additional molecular information that includes primer sequences for linkage analysis and quantitative measurement of knockdown. (D) A link to request the line.
Figure 2.
Example GBT expression summary—GBT0141. When image cataloging is completed for a GBT line, an expression summary is placed below the summary block. The expression summary is a grid of flattened z-stack images, specifically maximal image projections of a z-stack. The images from 2 (2DPF) and 4 (4DPF) days post-fertilization are arranged in vertical columns with the dorsal (D), sagittal (S) and ventral (V) images arranged in rows. This allows for quick viewing of the expression pattern, facilitating review of GBT lines for mRFP patterns of interest. By simultaneously viewing and comparing expression in different planes, some reconstruction of the three-dimensional expression pattern is possible.
mRFP expression patterns
Each mRFP-expressing GBT line represents a unique transposon insertion into a translated transcription unit. For many lines, an N-terminal fusion to mRFP will reflect the expression pattern of the endogenous gene. In a subset of fish, the mRFP fusion protein is secreted, and the mRFP expression reflects accumulation of the mRFP fusion protein due to non-autonomous biological systems or pathways (e.g. in filtering kidney, phagocytic blood or extracellular matrix). This dynamic mRFP expression is a key data deliverable of the GBT lines. At a minimum, GBT lines are currently imaged to capture mRFP expression at both 2 and 4 DPF, including dorsal, sagittal and ventral vantage points using SCORE imaging (7). The maximal image projection of a z-stack of images is stored within a media library for each line. Any multimedia files beyond the catalog images at 2 and 4 DPF can also be stored within the media gallery associated with each line. The media gallery is capable of storing images or movies in multiple formats (.jpg, .tif, .gif, .png, .mov, .m4v, .mpg, .avi and .wmv).
ZFA tags
With the large amounts of multimedia data, methods to make these data searchable are essential. We are using the ZFA Ontology originally developed by the zebrafish community and ZFIN to annotate tissues in normal and mutant zebrafish (8,9). Proper annotation of an organism is unlikely to occur through the efforts of a few individuals. Instead, we employ a community approach to leverage each individual's knowledge base to annotate tissues of interest. We developed an image annotation tool within zfishbook. A rectangular image tag can be resized and positioned to best mark mRFP-positive tissue, which is then annotated using ZFA terms. If the user is unfamiliar with ZFA terms, a link brings them to a search and lookup page. Once the ZFA term is entered, all parent tissues are also labeled. For example, in Figure 3 we show five image tags. When the mouse ‘hovers over’ an image tag it turns from white to yellow and the annotation appears (Figure 3, eyes → [head; visual_system]). Each tag and its parents are added to the images keywords and to the ZFA-annotated expression tags found within the summary block.
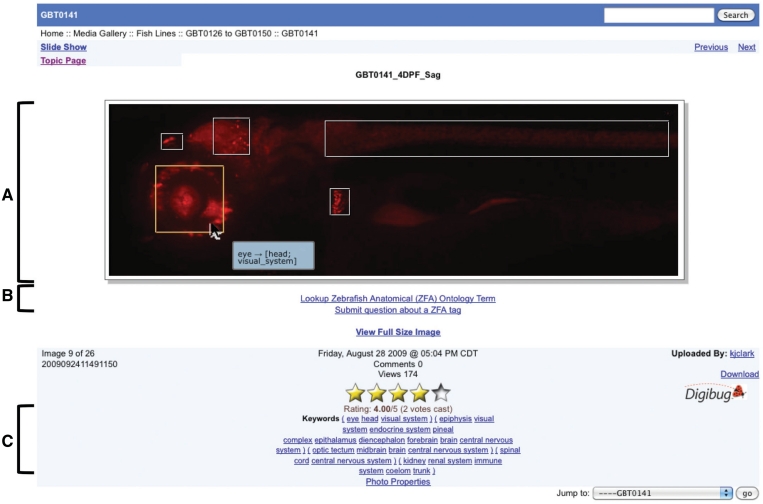
Figure 3.
Example of ZFA-tagged image—GBT0141, 4DPF, Sagittal. (A) This image has been tagged five times with our ZFA image tagging tool. Although, tagging requires a user login, viewing annotations is available to both unregistered and registered users. When viewing an image in the media gallery, the image tags appear as white rectangles over an mRFP expression domain. When hovering over the tag with the cursor, the rectangle turns yellow and the image tag is displayed. In this example, the cursor is hovering over the image tag over the eye that is part of both the head and the visual system. (B) A direct link to the ZFA lookup tool on zfishbook is below every image, as well as a link to send an email to question the annotation of the image. Only ZFA terms can be used to annotate images on zfishbook. (C) All the ZFA image tags become part of the image's keywords.
Molecular annotation
Disrupted genes are determined by multiple methods including cloning of transposon junction fragments from genomic DNA and identifying mRNA fusions to mRFP or GFP (5′- and 3′-Rapid Amplification of cDNA Ends (RACE), respectively). These sequence tags are stored within zfishbook and are available from the summary block as 5′ genomic, 3′ genomic, 5′-RACE or 3′-RACE categories. The 5′ and 3′ labels refer to orientation relative to the gene-break transposon since its mutagenic functionality is dependent on insertion orientation. Gene information is added to the summary block including its official symbol and name as well as alternate names as annotated in Entrez Gene (10). Gene links to external databases including Entrez Gene (10), Ensembl (11) and ZFIN (9) are included to facilitate molecular data retrieval by users. Candidate genes are then confirmed through linkage analysis to the observed mRFP pattern. The status of this process (candidate/confirmed) is listed in the summary block.
USING ZFISHBOOK
An online tutorial (http://zfishbook.org/tutorial) is available to help new users have a productive experience at zfishbook. In addition to explaining how to get to an individual line using the ‘Lines’ menu item, the tutorial helps new users discover lines of interest. One approach is to examine the lines with identified genes. Users can quickly find ‘Tagged Genes’ as a submenu of the ‘Lines’ menu. Once there, they can sort by date of identification, gene name, GBT line number or status (candidate or confirmed). A second approach is to examine expression patterns for lines of interest. As a new user, the best way to browse patterns is to look at the expression summary for each line (Figure 2) and view lines using the ‘Next Line’ link. Once a user is familiar with the lines, keeping up with new images can be done on a weekly or monthly basis using the ‘Recently Added Images’ within the ‘Media Gallery’ menu. As lines are annotated using the ZFA image tag tool, users can search for these lines based on the ZFA annotation tags. Using the ‘Search Lines’ under the ‘Line Classes’ menu item returns all lines with matching ZFA tags.
USER INPUT/DYNAMIC SITE
The zfishbook site is designed to be a multiuser and dynamic database. User login is not required to view the data provided by members of the IZPTC. User logins are only required to add data to the database. User login provides microannotation of contributions by those scientists creating new lines, adding images, molecular data, ZFA image tags or contributing to the digital notebook of a line. The data is placed directly onto zfishbook.org by individual users without quarantine. If labs outside of the IZPTC wish to perform screens using gene-break transposon technology, we are encouraging them to contribute this data to zfishbook.
DESIGN AND IMPLEMENTATION
zfishbook is implemented with a combination of open source Internet technologies. The open source MySQL (http://www.mysql.com/) relational database management system securely stores all data. Customized Hypertext Preprocessor (http://www.php.net/) server-side scripts interface with the database and generate HTML and JavaScript for the user's web browser to process. The open source glFusion (www.glfusion.org) content management system is used in conjunction with these technologies to provide advanced user and content management. Image tagging functionality is provided by the Tipmage JavaScript class (http://digitalhymn.com/argilla/tipmage/).
ACCESS
zfishbook can be freely accessed via the following methods:
| HTTP access: | http://www.zfishbook.org/ |
| Site updates RSS feed: | http://www.zfishbook.org/backend/updates.rss |
| Tagged gene list RSS feed: | http://www.zfishbook.org/backend/taggedGenes.rss |
| Database output flat file: | http://www.zfishbook.org/backend/flatfile.txt |
| Email access: | info@zfishbook.org |
FUTURE PLANS
GBT line generation is rapidly expanding, with multiple locations around the world generating novel fish lines for the broader scientific community. zfishbook represents a key central hub for the collection of this diverse data in a uniform format to allow community-wide contribution to line annotation.
FUNDING
National Institutes of Health, (grants DA14546, GM63904, HG 006431, NIDDK P30DK084567 for zfishbook.org to S.C.E., in part); Mayo Foundation. Funding for open access charge: GM63904.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank all members of the IZPTC, past and present, for their insight and helpful suggestions.
REFERENCES
- 1.Davidson AE, Balciunas D, Mohn D, Shaffer J, Hermanson S, Sivasubbu S, Cliff MP, Hackett PB, Ekker SC. Efficient gene delivery and gene expression in zebrafish using the Sleeping Beauty transposon. Dev. Biol. 2003;263:191–202. doi: 10.1016/j.ydbio.2003.07.013. [DOI] [PubMed] [Google Scholar]
- 2.Kawakami K, Takeda H, Kawakami N, Kobayashi M, Matsuda N, Mishina M. A transposon-mediated gene trap approach identifies developmentally regulated genes in zebrafish. Dev. Cell. 2004;7:133–144. doi: 10.1016/j.devcel.2004.06.005. [DOI] [PubMed] [Google Scholar]
- 3.Emelyanov A, Gao Y, Naqvi NI, Parinov S. Trans-kingdom transposition of the maize dissociation element. Genetics. 2006;174:1095–1104. doi: 10.1534/genetics.106.061184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sivasubbu S, Balciunas D, Davidson AE, Pickart MA, Hermanson SB, Wangensteen KJ, Wolbrink DC, Ekker SC. Gene-breaking transposon mutagenesis reveals an essential role for histone H2afza in zebrafish larval development. Mech. Dev. 2006;123:513–529. doi: 10.1016/j.mod.2006.06.002. [DOI] [PubMed] [Google Scholar]
- 5.Petzold AM, Balciunas D, Sivasubbu S, Clark KJ, Bedell VM, Westcot SE, Myers SR, Moulder GL, Thomas MJ, Ekker SC. Nicotine response genetics in the zebrafish. Proc. Natl Acad. Sci. USA. 2009;106:18662–18667. doi: 10.1073/pnas.0908247106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clark KJ, Balciunas D, Pogoda H-M, Ding Y, Westcot SE, Bedell VM, Greenwood TM, Urban MD, Skuster KJ, Petzold AM, et al. In vivo protein trapping produces a functional expression codex of the vertebrate proteome. Nat. Meth. 2011;8:506–512. doi: 10.1038/nmeth.1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Petzold AM, Bedell VM, Boczek NJ, Essner JJ, Balciunas D, Clark KJ, Ekker SC. SCORE imaging: specimen in a corrected optical rotational enclosure. Zebrafish. 2010;7:149–154. doi: 10.1089/zeb.2010.0660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sprague J, Bayraktaroglu L, Clements D, Conlin T, Fashena D, Frazer K, Haendel M, Howe DG, Mani P, Ramachandran S, et al. The Zebrafish Information Network: the zebrafish model organism database. Nucleic Acids Res. 2006;34:D581–D585. doi: 10.1093/nar/gkj086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bradford Y, Conlin T, Dunn N, Fashena D, Frazer K, Howe DG, Knight J, Mani P, Martin R, Moxon SAT, et al. ZFIN: enhancements and updates to the Zebrafish Model Organism Database. Nucleic Acids Res. 2011;39:D822–D829. doi: 10.1093/nar/gkq1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maglott D, Ostell J, Pruitt KD, Tatusova T. Entrez Gene: gene-centered information at NCBI. Nucleic Acids Res. 2010;39:D52–D57. doi: 10.1093/nar/gkq1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Flicek P, Amode MR, Barrell D, Beal K, Brent S, Chen Y, Clapham P, Coates G, Fairley S, Fitzgerald S, et al. Ensembl 2011. Nucleic Acids Res. 2011;39:D800–D806. doi: 10.1093/nar/gkq1064. [DOI] [PMC free article] [PubMed] [Google Scholar]