Abstract
Analogous to human leukocyte antigens, blood group antigens are surface markers on the erythrocyte cell membrane whose structures differ among individuals and which can be serologically identified. The Blood Group Antigen Gene Mutation Database (BGMUT) is an online repository of allelic variations in genes that determine the antigens of various human blood group systems. The database is manually curated with allelic information collated from scientific literature and from direct submissions from research laboratories. Currently, the database documents sequence variations of a total of 1251 alleles of all 40 gene loci that together are known to affect antigens of 30 human blood group systems. When available, information on the geographic or ethnic prevalence of an allele is also provided. The BGMUT website also has general information on the human blood group systems and the genes responsible for them. BGMUT is a part of the dbRBC resource of the National Center for Biotechnology Information, USA, and is available online at http://www.ncbi.nlm.nih.gov/projects/gv/rbc/xslcgi.fcgi?cmd=bgmut. The database should be of use to members of the transfusion medicine community, those interested in studies of genetic variation and related topics such as human migrations, and students as well as members of the general public.
INTRODUCTION
The BGMUT Blood Group Antigen Gene Mutation Database documents variations in genes that encode antigens for human blood groups. It is a part of the dbRBC resource (1) of the National Center for Biotechnology Information (NCBI) of USA and can be freely accessed online at http://www.ncbi.nlm.nih.gov/projects/gv/rbc/xslcgi.fcgi?cmd=bgmut.
Recent documentation of the extent and the surprisingly high numbers of mutations in the human genome have suggested that, perhaps with the exception of identical twins, no two individuals bear exact copies of chromosomal DNA. In those studies, DNA of random subjects is compared but more often phenotypic differences observed in disease states, whether single gene inherited disorders or in association studies of complex conditions are taken as criteria for selection of individuals whose DNA is examined for sequence changes. In the latter studies, large fragments of DNA are usually examined and compared statistically to matched control individuals. Changes in blood group phenotypes are another criterion for selection of subjects who may show differences in sequences of two or more defined sets of genes. These genes encode a group of red cell membrane proteins that are polymorphic in the population and are defined as blood group antigens; in addition, these genes may encode certain glycosyltransferases that are involved in the synthesis of red cell membrane glycans whose structures also differ among individuals. The former group consists of structural molecules, channels, adhesion molecules or enzymes and, excluding their red cell membrane location, can be considered as representative of any human protein, whereas the latter are similar to most other glycosyltransferases. Evidence suggests that sequence changes in these proteins or even their absence, such as in null phenotypes, is not, in most cases, physiologically harmful.
The proteins or the glycans fulfill their role as blood group antigens because they are polymorphic in the population and their sequence changes can be readily predicted by serological approaches; they are known as ‘antigens’ because in the course of transfusion, or pregnancy, the presence of a variant protein epitope is recognized as ‘non-self’ and may ultimately result in an adverse immunological reaction. The use of transfusion being ubiquitous in the practice of medicine, populations worldwide are serologically tested and variant antigens and their genes, in contrast to many other variant genes are being documented in a large number of diverse populations. Although some variants occur rarely and, may only be observed in a single individual or family, others appear in unexpectedly large populations, such as the MiIII phenotype encoded by the MiIII GYPA gene, in Taiwan (incidence can be as high as 88% among Ami tribes) (2). For many alleles, the database also provides information on the geographic or ethnic origin of alleles and/or their associated serological phenotypes when such information was presented in the publications describing the alleles. This may be of use to those interested in population migrations.
HISTORY AND CURRENT STATE
BGMUT was developed in 1999 as a locus-specific gene mutation database under the aegis of the Human Variation Genome Society. It was curated under the direction of one of the authors (OOB) with original information contributed by more than a dozen blood group system experts. The database was hosted online by the Department of Biochemistry at the Albert Einstein College of Medicine, New York. BGMUT was identified as one of three model locus-specific databases from more than 200 in a scholarly review (3). In 2006, BGMUT became a part of the dbRBC resource of the NCBI. At dbRBC, curatorship and direction for maintenance of the database has been provided by another of the authors (WH). The number of alleles in BGMUT has approximately doubled since 2004 when the database was first described in a scholarly publication (4). This publication has been referred to many times in the scientific literature indicating that BGMUT has been a useful resource. Links to BGMUT database records are available on relevant pages of the Wikipedia online encyclopedia, and on many of NCBI's online resources. BGMUT is also a part of the PhenCode project which attempts to integrate genetic variation data with the UCSC Genome Browser (5).
As of August 2011, BGMUT had 1251 alleles belonging to 40 genes that are together responsible for 30 human blood group systems (Table 1). Alleles of some genes, such as ABO and RHCE/D which are, respectively, responsible for the ABO and Rh blood group systems most frequently examined in the populations, are more numerous than those of others (Table 1). As per the International Society for Blood Transfusion, there are 30 human blood group systems (http://ibgrl.blood.co.uk/ISBT%20Pages/ISBT%20Terminology%20Pages/Table%20of%20blood%20group%20systems.htm), all of which are covered by BGMUT. The Globoside (GLOB) system is currently considered a part of the P1PK system in BGMUT, which additionally considers the system of T and Tn antigens as a separate blood group system.
Table 1.
Blood group systems in the BGMUT databasea
| System (symbol) | Genes |
|||
|---|---|---|---|---|
| Symbol | Chromosomal location | Nature of encoded protein | Alleles | |
| ABO (ABO) | ABO | 9q34.2 | Galactosyltransferase and N-acetylgalactosaminyltransferase | 272 |
| Chido/Rodgers | C4A | 6p21.3 | Complement factor | 3 |
| (CH/RG) | C4B | 6p21.3 | Complement factor | 4 |
| Colton (CO) | AQP1 | 7p14 | Water channel | 11 |
| Cromer (CROM) | CD55 | 1q32 | Complement cascade regulator | 15 |
| Diego (DI) | SLC4A1 | 17q21–q22 | Anion exchanger | 91 |
| Dombrock (DO) | ART4 | 12p13–p12 | ADP ribosyltransferase | 20 |
| Duffy (FY) | DARC | 1q21–q22 | Chemokine receptor | 11 |
| Gerbich (GE) | GYPC | 2q14–q21 | Cytokeletal element | 10 |
| Gill (GIL) | AQP3 | 9p13 | Water channel | 8 |
| H (H) | FUT1 | 19q13.3 | Fucosyltransferase | 45 |
| FUT2 | 19q13.3 | Fucosyltransferase | 56 | |
| I (I) | GCNT2 | 6p24.2 | N-acetylglucosaminyltransferase | 13 |
| Indian (IN) | CD44 | 11p13 | Adhesion molecule | 4 |
| John Milton Hagen (JMH) | SEMA7A | 15q22.3–q23 | Adhesion and signaling molecule? | 12 |
| Kell (KEL) | KEL | 7q33 | Metalloendopeptidase? | 57 |
| Kidd (JK) | SLC14A1 | 18q11–q12 | Urea transporter | 18 |
| Knops (KN) | CR1 | 1q32 | Cell surface receptor | 32 |
| Kx (XK) | XK | Xp21.1 | Membrane transport protein? | 35 |
| Landsteiner-Weiner (LW) | ICAM4 | 19p13.2-cen | Adhesion molecule | 4 |
| Lewis (LE)b | FUT3 | 19p13.3 | Fucosyltransferase | 52 |
| FUT6 | 19p13.3 | Fucosyltransferase | 20 | |
| FUT7 | 9q34.3 | Fucosyltransferase | 2 | |
| Lutheran (LU) | BCAM | 19q13.2 | Adhesion molecule | 20 |
| MNS (MNS) | GYPA | 4q31.21 | Unknown | 38c |
| GYPB | 4q31.21 | Unknown | 35c | |
| GYPE | 4q31.1 | Unknown | 2c | |
| Ok (OK) | BSG | 19p13.3 | Plasma membrane protein | 5 |
| P1PK (P1PK) including globoside (GLOB) | B3GALNT1 | 3q25 | N-acetylgalactosaminyltransferase | 9 |
| A4GALT | 22q13.2 | Galactosyltransferase | 37 | |
| Raph (RAPH) | CD151 | 11p15.5 | Adhesion molecule | 4 |
| Rh (RH) | RHCE | 1p36.11 | Unknown | 103 |
| RHD | 1p36.11 | Unknown | 185 | |
| Rh-associated glycoprotein (RHAG) | RHAG | 6p21.1–p11 | Ammonium and carbon dioxide transporter | 23 |
| Scianna (SC) | ERMAP | 1p34.2 | Adhesion molecule? | 9 |
| T/Tn | C1GALT1C1 | Xq24 | Galactosyltransferase | 8 |
| C1GALT1 | 7p21.3 | Galactosyltransferase | 1 | |
| Xg (XG) | XG | Xp22.33 | Unknown | 1 |
| CD99 | Xp22.32, Yp11.3 | Adhesion molecule | 1 | |
| Yt (YT) | ACHE | 7q22 | Acetylcholinesterase | 4 |
aAs of August 2011; total 30 systems, 40 genes and 1251 alleles.
bThe FUT3, FUT6 and FUT7 genes are not expressed in erythroid cells.
cIncluding hybrid alleles with another member of the GYPA/B/E gene family.
DATABASE ARCHITECTURE
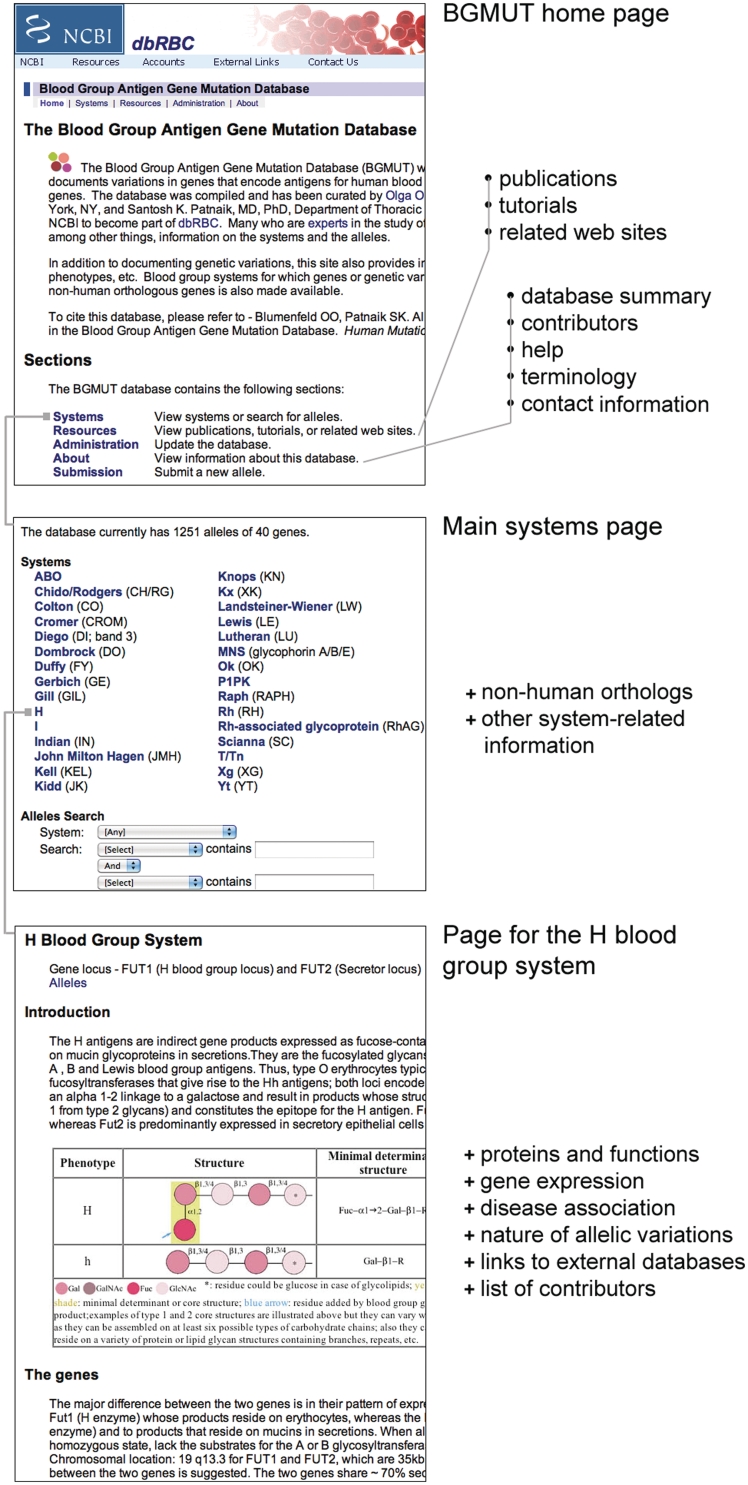
BGMUT is accessible online for view, search or for administration as a website in the form of HTML (hyper-text markup language) pages (Figures 1 and 2). A Microsoft SQL Server relational database is used for data storage. Programmatic code in SQL (structured query language), C++ and XSLT (extensible structured language transformations) languages is used for interaction with the SQL Server database and for rendering BGMUT's web-based interface. Raw data on all or a subset of the alleles described in BGMUT can be downloaded as tab-delimited or comma-separated (CSV) text formats from the BGMUT website. Compilations of allelic sequences for the ABO, H, MNS and Rh systems in the Microsoft Excel format are also available for download.
Figure 1.
A montage of screenshots of the web-based interface of BGMUT depicting the hierarchically ordered home page, the page listing the blood group systems in the database and the page for a specific blood group system (H). The nature of content of the pages is visible in the screenshots or is noted in the figure.
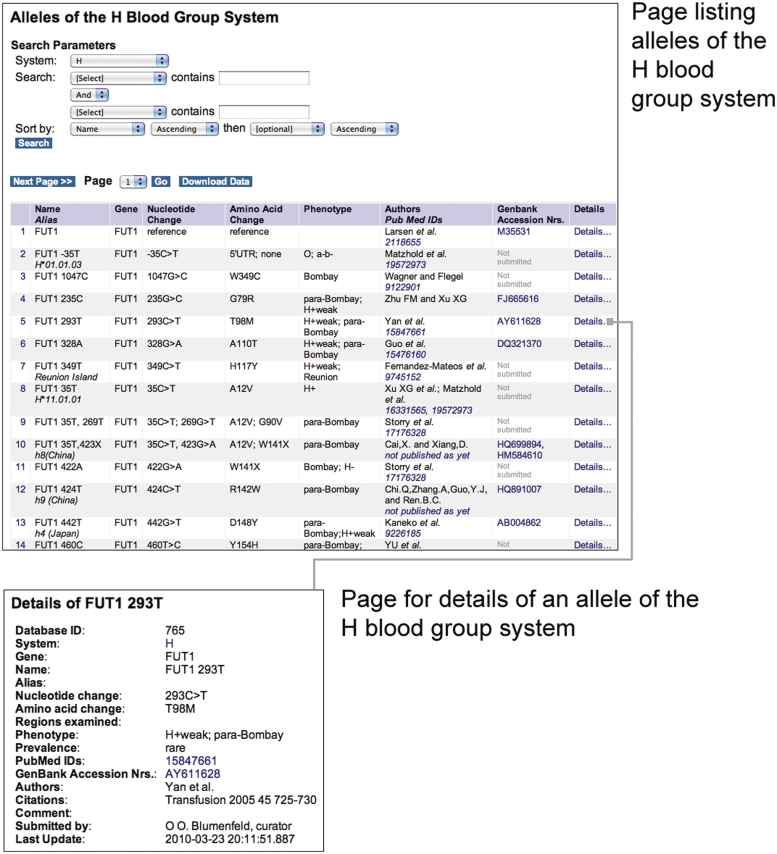
Figure 2.
Screenshots of the BGMUT web pages showing a partial list of alleles and a specific allele of the H blood group system.
DATABASE CURATION AND ALLELE SUBMISSION
The BGMUT database is curated manually. Allelic information is periodically collated from scientific literature, as is the case for a majority of the alleles listed in BGMUT, or is obtained as direct submissions from researchers through the database website. During the process of curation, good quality of methods used in a study is ascertained. The new candidate allele's sequence that has been published and/or submitted to a publicly available repository such as NCBI's GenBank is compared to BGMUT's reference allele for the gene sequence. The sequence positions and the kinds of the deduced amino acid changes are also verified. Authors are consulted in case of a question or disagreement. For direct submission of information on a new allele for inclusion in the database, a scientific publication describing the allele is not required. However, submitters are encouraged to deposit the allele's sequence in a publicly available repository. In the absence of a scientific publication, this is a requirement.
ALLELES IN BGMUT
Alleles in BGMUT are grouped by the blood group system that the genes they belong to affect. For each allele in the database, BGMUT provides details on the nucleotide changes and the deduced amino acid changes in the protein encoded by the gene the allele belongs to. These changes are in context of a ‘reference’ allele that itself is included in BGMUT, and is the same for all alleles of a gene. Besides the information on the sequence changes, BGMUT also details for an allele the frequency of occurrence, the associated blood group phenotype, references to the studies that identified and characterized the allele and accession numbers of the relevant sequences in NCBI GenBank when such information is available. GenBank accession numbers, however, are not available for many alleles because though they have been described in published literature, their sequences were not deposited in the repository by the authors. When known, the regions of the gene or cDNA that were sequenced to identify the allele, the prevalence of the allele in different geographical regions or ethnic populations and association of the allele with diseases are also noted.
Often, a name is also provided for an allele. Names make it easier to refer to alleles and can indicate the associated phenotype and/or nucleotide or amino acid variation. BGMUT uses the allele names given by the discoverers of the alleles and/or those that are commonly used by blood group system experts. In the absence of such a name, the curator may provide an arbitrary name. It should be noted that every allele entry in BGMUT has a unique database identification number associated with it.
The naming of alleles has presented a problem compounded by names given to serological phenotypes of antigens they encode. At this time, efforts by the transfusion medicine community are in progress to standardize and simplify this nomenclature (6). In the database, to more generally identify each allele, in most cases, ‘name’ includes the gene name followed by the DNA change particular to the allele; to facilitate the recognition of the phenotype//genotype connection, under ‘alias’ is given the serological designation as used by the original authors. In some cases, such as for the Rh system, because of traditional reasons, this arrangement is reversed; in others, such as ABO, only the phenotype designation marked by consecutive numbers is given; this is because the large number of sites of DNA changes in many alleles makes their listing under ‘name’ unwieldy.
Alignments of DNA changes in alleles of a single gene or a gene family may uncover patterns that provide some insights into the process of diversification, including gene rearrangements and meiotic or unequal recombination (7). An attempt to correlate DNA changes in the gene(s) encoding the antigen(s), with the serological phenotype or the glycosyltransferase activity, shows that even though variation in common alleles occurs within the same sequence stretches that define the common epitopic regions (4), in rare alleles, changes that affect the serological response are seen throughout the exons. The resulting amino acid alterations may cause altered folding that interferes with epitope presentation or gives rise to new epitopes. As expected, null phenotypes usually result from absence of an intact protein and are caused by nonsense mutations, insertions or deletions, or absence of the common epitope due to recombination. More generally, in the systems documented in the database, the phenotype reflects a particular sequence alteration be it the result of a single or multiple molecular events (e.g. the Rh neg, ABO O and MNS Sta alleles). Clearly, in contrast to many global studies, data presented in BGMUT allow to focus on DNA changes in a single gene and a closer look at its resulting alleles, to ask how they came about and to consider the properties of the resulting proteins. This knowledge will only expand as complete sequence content of variant alleles become more readily available.
OTHER RESOURCES WITHIN BGMUT
Besides the alleles of 30 human blood group systems, BGMUT also provides information on the systems themselves (Figure 1). Such information includes that on a system's genes, such as their sequences, chromosomal locations and the functions of proteins encoded by them. Links to database records elsewhere, such as the Online Mendelian Inheritance in Man (OMIM) (8) and PubMed resources of NCBI, are provided. Brief descriptions of diseases associated with a gene's variations, if any, and a summary of the nature of the variations are also provided. Content for such pages for the various blood group systems has been contributed by various experts and is continually updated. The BGMUT website also has a few introductory tutorials on blood group systems and provides links to online resources on blood groups as well as on human gene mutations and polymorphisms in general. Information on the nature of the genes of the H, MNS, Rh and RhAG systems in non-humans, primarily primates and their allelic variations is also provided. BGMUT also makes available brief descriptions of other carbohydrate antigens such as the Tk and Sid (Sda) antigens on erythrocytes.
ADDITIONAL TOOLS OF INTEREST AT THE PARENT dbRBC RESOURCE
Besides BGMUT, the dbRBC resource contains tools and other resources developed at the NCBI to provide access to publicly available information on human blood group systems. Two of the tools are briefly described here. The alignment viewer tool (http://www.ncbi.nlm.nih.gov/projects/gv/rbc/align.fcgi?cmd=aligndisplay) provides an interface to choose alleles of genes of many of the blood group systems for sequence alignment, though allelic entries in BGMUT cannot be directly chosen in the current interface. Chosen sequences can be displayed as genomic DNA, cDNA or amino acid sequences. Positions known to bear gene polymorphisms can be highlighted, and so can be specific differences within a set of allelic sequences. The reference sequence itself can be chosen. Besides the alignment, a user can switch to a display of selected sequences in FASTA format. Nucleotide or protein sequences for individual exons and/or introns can be displayed for each allele in alignment to the reference sequence. When the three dimensional structure of a gene's protein product is known, amino acid differences of various alleles can be projected onto the protein model for viewing with the CN3D model viewer (9).
dbRBC's sequencing-based typing tool (http://www.ncbi.nlm.nih.gov/projects/gv/rbc/sbt.cgi?cmd=main) is of use to evaluate the allelic composition of sequencing-based typing results of cDNA or genomic sequences (10). It allows for the comparison of result sequences with sequences of known alleles of genes of various human blood group systems. The results of the comparison are returned as a table of potential allele hits, along with the respective nucleotide changes, and links for closer examination of the alignments within the dbRBC alignment viewer tool.
With a continuous increase in the number of available alleles, dbRBC is testing the implementation of an allelic nomenclature system based on one in use for the HLA system (11). Thus, for example, the reference ABO gene allele sequence is named ABO*A1.01.01.01. The name consists of the gene name, followed by an asterisk, and for blocks separated by periods. The leading block is to group sequences first by their main serological phenotypes (e.g. A1, A2 or Ax phenotypes of the ABO system). Sequences that share the same serological phenotype, but differ in their amino acid sequences, are differentiated by the second block. The third block lists alleles that have an identical amino acid sequence, but show synonymous mutations in their exonic DNA sequence. The last block differentiates sequences that share an identical DNA sequence in the coding region, but differ in non-coding regions. Alleles of the ABO, FUT1, FUT2 and FUT3 genes have been given additional names as per this system as of July 2011.
FUTURE PLANS
Currently, allelic information compiled in BGMUT is not integrated with that outside the BGMUT but within the dbRBC resource. The latter set of alleles of genes for human blood group systems have been programmatically collected from various NCBI resources. Plans for an integration so that BGMUT allelic entries are available for use within the alignment viewer and the sequencing-based typing tools of dbRBC are under consideration. As a part of this integration, BGMUT alleles will be assigned additional names as per the nomenclature system outlined in the above section if a decision is made to implement it after consultation with various experts.
FUNDING
Funding for developing and maintaining the Database, was provided by the Department of Biochemistry, Albert Einstein College of Medicine of Yeshiva University, N.Y., 10461, USA, and from the David Opochinsky/Blumenfeld Family Fund (to O.O.B.). Intramural funding of the US National Institutes of Health (to NCBI and W.H.)
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank Raymond Dunivin and Douglas Hoffman at NCBI for help with the implementation and maintenance of the BGMUT database.
REFERENCES
- 1.Sayers EW, Barrett T, Benson DA, Bolton E, Bryant SH, Canese K, Chetvernin V, Church DM, DiCuccio M, Federhen S, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2011;39:D38–D51. doi: 10.1093/nar/gkq1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Broadberry RE, Lin M. The distribution of the MiIII (Gp.Mur) phenotype among the population of Taiwan. Transfus. Med. 1996;6:145–148. doi: 10.1046/j.1365-3148.1996.d01-64.x. [DOI] [PubMed] [Google Scholar]
- 3.Claustres M, Horaitis O, Vanevski M, Cotton RG. Time for a unified system of mutation description and reporting: a review of locus-specific mutation databases. Genome Res. 2002;12:680–688. doi: 10.1101/gr.217702. [DOI] [PubMed] [Google Scholar]
- 4.Blumenfeld OO, Patnaik SK. Allelic genes of blood group antigens: a source of human mutations and cSNPs documented in the Blood Group Antigen Gene Mutation Database. Hum. Mutat. 2004;23:8–16. doi: 10.1002/humu.10296. [DOI] [PubMed] [Google Scholar]
- 5.Giardine B, Riemer C, Hefferon T, Thomas D, Hsu F, Zielenski J, Sang Y, Elnitski L, Cutting G, Trumbower H, et al. PhenCode: connecting ENCODE data with mutations and phenotype. Hum. Mutat. 2007;28:554–562. doi: 10.1002/humu.20484. [DOI] [PubMed] [Google Scholar]
- 6.Storry JR, Castilho L, Daniels G, Flegel WA, Garratty G, Francis CL, Moulds JM, Moulds JJ, Olsson ML, Poole J, et al. International Society of Blood Transfusion Working Party on red cell immunogenetics and blood group terminology: Berlin report. Vox Sang. 2011;101:77–82. doi: 10.1111/j.1423-0410.2010.01462.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Patnaik SK, Blumenfeld OO. Patterns of human genetic variation inferred from comparative analysis of allelic mutations in blood group antigen genes. Hum. Mutat. 2011;32:263–271. doi: 10.1002/humu.21430. [DOI] [PubMed] [Google Scholar]
- 8.Hamosh A, Scott AF, Amberger JS, Bocchini CA, McKusick VA. Online Mendelian Inheritance in Man (OMIM), a knowledgebase of human genes and genetic disorders. Nucleic Acids Res. 2005;33:D514–D517. doi: 10.1093/nar/gki033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang Y, Geer LY, Chappey C, Kans JA, Bryant SH. Cn3D: sequence and structure views for Entrez. Trends Biochem. Sci. 2000;25:300–302. doi: 10.1016/s0968-0004(00)01561-9. [DOI] [PubMed] [Google Scholar]
- 10.Helmberg W, Dunivin R, Feolo M. The sequencing-based typing tool of dbMHC: typing highly polymorphic gene sequences. Nucleic Acids Res. 2004;32:W173–W175. doi: 10.1093/nar/gkh424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Robinson J, Mistry K, McWilliam H, Lopez R, Parham P, Marsh SG. The IMGT/HLA database. Nucleic Acids Res. 2011;39:D1171–D1176. doi: 10.1093/nar/gkq998. [DOI] [PMC free article] [PubMed] [Google Scholar]