Abstract
Nucleic acid phylogenetic profiling (NAPP) classifies coding and non-coding sequences in a genome according to their pattern of conservation across other genomes. This procedure efficiently distinguishes clusters of functional non-coding elements in bacteria, particularly small RNAs and cis-regulatory RNAs, from other conserved sequences. In contrast to other non-coding RNA detection pipelines, NAPP does not require the presence of conserved RNA secondary structure and therefore is likely to identify previously undetected RNA genes or elements. Furthermore, as NAPP clusters contain both coding and non-coding sequences with similar occurrence profiles, they can be analyzed under a functional perspective. We recently improved the NAPP pipeline and applied it to a collection of 949 bacterial and 68 archaeal species. The database and web interface available at http://napp.u-psud.fr/ enable detailed analysis of NAPP clusters enriched in non-coding RNAs, graphical display of phylogenetic profiles, visualization of predicted RNAs in their genome context and extraction of predicted RNAs for use with genome browsers or other software.
INTRODUCTION
In all living organisms, the non-protein-coding regions of genomes are host to a high density of functional elements, including regulatory DNA sequences, transcription terminators and attenuators, riboswitches and wide variety of non-coding RNA genes such as SRP RNA, RNAse P RNA or regulatory RNAs. Most bacterial genomes are thought to harbor in the order of a hundred small regulatory RNAs (sRNAs) and a larger number of riboswitches, T-boxes and regulatory leader elements located in the 5′ regions of mRNA genes (cis-encoded RNAs). Traditionally, genome annotation processes have focused mostly on protein-coding genes and ignored these elements, to the point that the notion of ‘gene’ was (and still is) confused with that of ‘CDS’ (coding sequence) in most bacterial and archaeal genome annotations. These ‘white spaces’ between coding sequences are now being intensely scrutinized for regulatory elements and non-coding RNA (ncRNA) genes. Early ncRNA gene finders used the higher GC contents and supposed propensity for secondary structure formation of RNAs to identify potential ncRNA genes (1). However, as soon as complete genomes were available in sufficient numbers, these approaches were superseded by comparative genomics, which proved much more powerful in discriminating functional elements from ‘junk’ DNA (1). Comparative genomics reveals thousands of conserved elements in the intergenic regions of even small bacterial genomes. However, these elements are not always ncRNAs and biologists need specific computational tools to discriminate ncRNAs from other classes of functional elements or noise. The first of these tools was qRNA (2) that uses pairwise sequence alignments to distinguish structural RNAs from protein-coding segments by seeking mutational patterns consistent with a base-paired secondary structure or with a coding sequence. Another program, RNAz (3), uses a multiple sequence alignment to classify conserved segments into ncRNA/non-ncRNA, based on the detection of significant conserved secondary structure and base-pair covariation. A more recent ncRNA detection procedure, sRNAPredict/SIPHT (4), was developed specifically for bacteria and combines intergenic sequence conservation to the detection of Rho-independent transcription terminators (RIT).
There is no gold standard for declaring a conserved non-coding sequence an RNA gene, a cis-encoded RNA, a DNA-level element or a mere sequence comparison artifact. Both qRNA and RNAz assume that functional ncRNAs adopt conserved secondary structures. However, some bacterial sRNAs may act independently of a specific secondary structure (5). The sRNAPredict pipeline requires that candidate ncRNA genes are flanked by a RIT. However, a significant fraction of bacteria prefer other termination mechanisms and, even in those that favor RITs, many genes use Rho-dependent termination or are part of operons and therefore are not followed directly by a terminator. There is thus a strong need for methods that can analyze all non-coding genome elements independently of associated secondary structures.
We have introduced nucleic acid phylogenetic profiling (NAPP) as a method for the classification of intergenic conserved non-coding elements (CNEs) in bacterial genomes (6). NAPP involves collecting all CNEs and CDSs in a reference species and seeking homologous sequences in all other available species. CNEs and CDSs are then clustered based on the similarity of their occurrence profiles. Applying this procedure to several bacterial genomes, we observed that actual ncRNA genes and cis-acting RNAs tend to strongly cluster together. We inferred that unidentified CNEs in these clusters were good candidates for RNA genes or elements, and experimentally confirmed this for a set of Staphylococcus aureus candidates. We since applied NAPP to the identification of RNA genes in Bacillus subtilis (7) and improved the pipeline in several respects (see below). To this date, however, distributing NAPP results to a wider community has proven difficult due to the relative complexity of NAPP outputs and the quickly expanding bacterial genome set. We have now developed a database and web server for distributing NAPP data. The current version of the database covers 1017 species including 949 Bacteria and 68 Archaea.
THE NAPP PIPELINE
A significant change to the NAPP pipeline since its first publication relates to the definition of CNEs. In the original NAPP procedure, we identified CNEs as segments of variable length with a minimal conservation ratio, computed from local Blast (8) alignments against other genomes. This presented two drawbacks: first, it discarded any region with conservation lower than a defined cutoff, which was problematic as we know many RNAs are poorly conserved. Second, the resulting CNEs possibly combined elements with different conservation profiles. We were interested in a more precise definition of CNEs that would handle domains with different evolutionary histories as independent entities. To address this, we developed a tiling procedure where each intergenic region of the reference genome is divided into 50 nt, non-oriented tiles overlapping by 25 nt.
All genes and intergenic tiles are submitted to the same clustering procedure. Here, we define as ‘gene’ any protein- or RNA-coding element provided in Genbank annotation files, i.e. protein-coding genes are restricted to their coding part and RNA genes are mostly tRNAs and rRNAs. We align each tile/gene from the reference organism against a set of 1069 bacterial/archaeal genomes (obtained from the NCBI server in mid-2010) using NCBI BLASTN 2.2.15 (parameters: −W 7 −e 0.01). The highest bit score obtained against each genome is normalized by the Blastn score of the sequence against itself. The phylogenetic profile of each tile/gene is thus a vector of 1069 normalized scores. Profiles are clustered using the K-means method (R package) with parameters: Pearson distance and k = 50. To identify ncRNA-rich clusters, we Blast all tiles in a cluster (parameters: −W 5 −e 0.001) against the RFAM-full 10.0 database (9). Tiles with Blast hits are tagged as ‘RNA’ tiles. Each cluster is assigned an ‘RNA enrichment P-value’ using Fisher's exact test (R package) based on counts of ‘RNA’ and ‘normal’ tiles. Clusters with a Fisher's P < 0.05 are considered as ‘RNA-rich’. We measure Gene Ontology (GO) term enrichments of RNA-rich clusters based on their protein-coding gene contents. We convert the GI numbers of protein-coding genes into Uniprot IDs and then into GO terms using the appropriate EBI conversion tables. This retrieves GO terms for 66% of CDSs. We measure terms enrichment in RNA-rich clusters using a Fisher's exact test.
The NAPP database is implemented using MySQL relational database server software version 4.1.22. The interface is developed using PHP5 version 4.3.9, hosted via an Apache server. Parts of the interface use Javascript. Tables and profile views are constructed on the fly using PHP and database queries.
RESULTS
RNA-rich clusters are found in 1017 of the 1069 genomes analyzed. Species that fail to produce RNA-rich clusters are mostly Archaea, endosymbionts (Buchnera, Carsonella and Mycoplasma) and other bacteria with unusually compact genomes (Supplementary Table S1), suggesting that these species contain fewer RNA genes and elements than average. There are in average 4.3 RNA-rich clusters per species containing altogether 1330 tiles. For ncRNA prediction purposes, we combine all tiles separated by <100 nt into contigs, independent of their cluster of origin (contigs are not oriented). This procedure produces an average of 643 contigs per genome, or 179 contigs/Mbase of genomic sequence (Supplementary Table S2). A large fraction of contigs (49.5%) are composed of a single tile (Supplementary Table S3). As could be intuitively expected, large contigs are more likely to represent actual RNAs than short ones. Contigs made of four tiles or more are four times as likely to match RFAM RNAs than single-tile contigs (corrected for size, Supplementary Table S3, last column). This suggests that prospective experimental validation should address contigs over four tiles in priority.
The ncRNA prediction accuracy of NAPP was recently benchmarked (10) along with the three other comparative genomics methods: sRNAPredict/SIPHT, RNAz and eQRNA. The benchmark used a test set of 776 sRNAs from 10 bacterial species, including 132 RNAs from RFAM and the others from RNA-seq and tiling array experiments. NAPP was generally more sensitive (higher recall rate) but less specific (more ‘false positive’ predictions) than other methods. As sRNAPredict had the best overall accuracy (weighting both sensitivity and specificity), we further compare NAPP with this method below.
NAPP predicts much more RNA loci than sRNAPredict. Averaged over all genomes, sRNAPredict finds 25 RNA loci/Mb (4) while NAPP finds 179 RNA loci/Mb (Supplementary Table S1). While this explains in part the higher reported specificity of sRNAPredict [12% versus 4% for NAPP (10)], it should be noted that specificity is notoriously difficult to evaluate in this case (10) as many false positives may turn out to be expressed in rare conditions. Furthermore, NAPP predictions include a significant number of ncRNAs lacking a RIT that are thus excluded by sRNAPredict. This comprises a large fraction of riboswitches and other mRNA leaders that use translational attenuation instead of premature termination, and most of the sRNA predictions in genomes with low RIT usage. RIT usage is low overall in Actinobacteria as well as in a number of isolated Spirochaetes, Cyanobacteria, Alphaproteobacteria or Firmicutes. These genomes were not sampled in the benchmark (10), with the exception of Caulobacter crescentus. We tested 21 genomes with a reportedly low RIT usage (11) and observed a dramatic decrease in the number of sRNAPredict loci for these genomes, from 25/Mb in average to 2/Mb in the low RIT set, while the number of NAPP predictions remains stable at 159/Mb versus 179/Mb in average (Supplementary Tables S4 and S5). Interestingly, these RIT-poor genomes have nearly no RNA annotated and few RFAM hits besides housekeeping RNAs and CRISPR loci, which suggests NAPP predictions may be a rich source of RNA discovery in these species. It should be noted that NAPP also predicts RNAs in Archaea (which do not contain RIT), at an average density of 142/Mb (Supplementary Table S2).
An underlying hypothesis in phylogenetic profiling is that genes or elements co-occur because they contribute to the same cellular function (12). We noted in our initial study that many ncRNAs tend to cluster with housekeeping genes. We later identified a cluster of B. subtilis elements enriched in sporulation genes and containing a novel sRNA gene expressed during sporulation (7). To enable this kind of analysis, we built GO term bias analysis into the NAPP server. The most frequent GO terms biases in RNA clusters (Supplementary Table S6) are related to housekeeping functions, such as translation (‘ribosome’, ‘translation’, ‘translational elongation’, etc.) and energy metabolism (‘proton transport’, ‘NADH dehydrogenase’, ‘ATP hydrolysis’, ‘ATP synthesis’, etc.). Terms related to RNA binding (‘RNA binding’, ‘ribonucleoprotein complex’, ‘rRNA binding’ and ‘tRNA binding’) are also very frequent. This enrichment is intriguing as it suggests a co-evolution of certain RNAs and RNA-binding proteins; however, it is more likely a consequence of an over-representation of RNA-binding activities among housekeeping functions. Another family of terms found in fewer species is ‘transposition’, ‘transposase’, etc. This is interesting as these terms are often associated with characteristic ‘patchy’ phylogenetic profiles, such as the one shown in Figure 1B (first column) for Escherichia coli strain O127 H6. These profiles are often associated to plasmid or transposon-borne elements that are horizontally transmitted across distant bacteria.
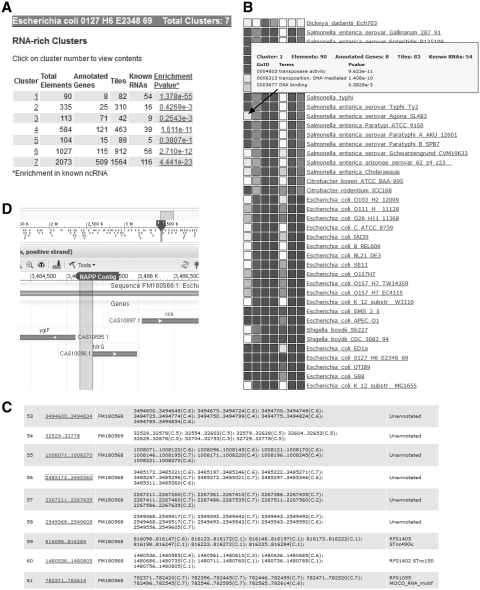
Figure 1.
Main displays of the NAPP database interface. (A) ‘RNA-rich Clusters’ table: for each cluster, the table provides cluster number, total number of elements, number of annotated genes, number of tiles, number of known ncRNAs (RFAM) and P-value of ncRNA enrichment. (B) Profile page: each column represents the average phylogenetic profile of all members of an RNA-rich cluster. There are as many lines as species in the database (at present 1017). Shaded/white squares indicate that members of this cluster have/do not have homologs in this species. Darkness indicates average Blast scores of homologues (darker: higher scores). The popup window displays GO term biases and other information on cluster #1. (C) Contig page. Contigs are produced by aggregating all overlapping or adjacent tiles from RNA-rich clusters. A single contig may contain tiles from different clusters. Columns indicate contig number, Genbank id. of chromosome, tiles forming contig (the cluster number of each tile is indicated in parentheses as C.1, C.2, etc.) and RFAM annotation. (D) Context view: from the contig or cluster views, users can visualize tiles or contigs in their genomic context through links to the NCBI genome server.
USING THE NAPP DATABASE
The NAPP database is freely available online at http://napp.u-psud.fr/. The interface is composed of the following display elements (further detail is provided in the Figure 1 legend and in the online documentation).
Main page: NAPP predictions are accessed for one species at a time. Species are selected either by typing keywords or through a drop-down menu.
‘RNA-rich clusters’ table (Figure 1A): this is the central page for a selected species. It presents the main characteristics of RNA-rich clusters. From this page, users can enter a position range to list the elements of interest (tiles and genes) at this location, or they can access the following pages.
Cluster view page: this page displays the contents of an RNA-rich cluster (element name, position, Id and description).
Profile page (Figure 1B): this page presents a graphical view of the phylogenetic profiles of RNA-rich clusters, together with associated GO-term biases.
Contig page (Figure 1C): this page displays contigs produced from adjacent tiles in all RNA-rich clusters.
Genome context view (Figure 1D): from the cluster and contig pages, users can visualize any element (tile, gene or contig) in its genomic context using the NCBI bacterial genome browser.
For each genome, users can export tiles or contigs as CSV or GFF files for further analysis or visualization in a genome browser. Queries and comments are welcomed at napp.biologie@u-psud.fr.
FUTURE DIRECTIONS
The long-term objective of the NAPP project is to provide biologists with comprehensive information on CNEs in bacterial and archaeal genomes. While the current implementation focuses on the subset of elements that co-evolve with known non-coding RNAs, many non-coding elements harbor distinct phylogenetic profiles that are not captured by this procedure. Furthermore, current NAPP clusters can be very large (up to several thousand tiles and genes) and may combine tiles and genes with slightly, albeit significantly, different profiles that would benefit from a more detailed examination. For instance, the ‘sporulation’ cluster, we identified in B. subtilis (7) is actually part of cluster #2 for this species that is enriched in housekeeping terms (‘ribosome’, ‘translation’, etc.) but not in the ‘sporulation’ term. The sporulation subcluster is visible only through visual inspection of a hierarchical clustering tree, which is not permitted with the current interface. A more comprehensive and interactive analysis of complete clustering trees should lead both to improved functional classification and to the recovery of additional non-coding elements that are currently discarded from NAPP clusters.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables S1–S6.
FUNDING
Agence Nationale pour la Recherche (grant ANR-2010-BLAN-1602-01) ‘Duplex-omics’. Funding for open access charge: Agence Nationale pour la Recherche, France.
Conflict of interest statement. None declared.
REFERENCES
- 1.Backofen R, Hess WR. Computational prediction of sRNAs and their targets in bacteria. RNA Biol. 2010;7:33–42. doi: 10.4161/rna.7.1.10655. [DOI] [PubMed] [Google Scholar]
- 2.Rivas E, Eddy SR. Noncoding RNA gene detection using comparative sequence analysis. BMC Bioinformatics. 2001;2:8. doi: 10.1186/1471-2105-2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Washietl S, Hofacker IL, Stadler PF. Fast and reliable prediction of noncoding RNAs. Proc. Natl Acad. Sci. USA. 2005;102:2454–2459. doi: 10.1073/pnas.0409169102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Livny J, Teonadi H, Livny M, Waldor MK. High-throughput, kingdom-wide prediction and annotation of bacterial non-coding RNAs. PLoS ONE. 2008;3:e3197. doi: 10.1371/journal.pone.0003197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Papenfort K, Bouvier M, Mika F, Sharma CM, Vogel J. Evidence for an autonomous 5' target recognition domain in an Hfq-associated small RNA. Proc. Natl Acad. Sci. USA. 2010;107:20435–20440. doi: 10.1073/pnas.1009784107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marchais A, Naville M, Bohn C, Bouloc P, Gautheret D. Single-pass classification of all noncoding sequences in a bacterial genome using phylogenetic profiles. Genome Res. 2009;19:1084–1092. doi: 10.1101/gr.089714.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marchais A, Duperrier S, Durand S, Gautheret D, Stragier P. CsfG, a family of sporulation-specific, small non-coding RNA highly conserved in endospore formers. RNA Biol. 2011;8:358–364. doi: 10.4161/rna.8.3.14998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 9.Gardner PP, Daub J, Tate JG, Nawrocki EP, Kolbe DL, Lindgreen S, Wilkinson AC, Finn RD, Griffiths-Jones S, Eddy SR, et al. Rfam: Updates to the RNA families database. Nucleic Acids Res. 2008;37:D136–D140. doi: 10.1093/nar/gkn766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lu X, Goodrich-Blair H, Tjaden B. Assessing computational tools for the discovery of small RNA genes in bacteria. RNA. 2011;17:1635–1647. doi: 10.1261/rna.2689811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Hoon MJ, Makita Y, Nakai K, Miyano S. Prediction of transcriptional terminators in Bacillus subtilis and related species. PLoS Comput. Biol. 2005;1:e25. doi: 10.1371/journal.pcbi.0010025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pellegrini M, Marcotte EM, Thompson MJ, Eisenberg D, Yeates TO. Assigning protein functions by comparative genome analysis: protein phylogenetic profiles. Proc. Natl Acad. Sci. USA. 1999;96:4285–4288. doi: 10.1073/pnas.96.8.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]