Abstract
Autism spectrum disorder (ASD) is a heterogeneous neurodevelopmental disorder with a prevalence of 0.9–2.6%. Twin studies showed a heritability of 38–90%, indicating strong genetic contributions. Yet it is unclear how many genes have been associated with ASD and how strong the evidence is. A comprehensive review and analysis of literature and data may bring a clearer big picture of autism genetics. We show that as many as 2193 genes, 2806 SNPs/VNTRs, 4544 copy number variations (CNVs) and 158 linkage regions have been associated with ASD by GWAS, genome-wide CNV studies, linkage analyses, low-scale genetic association studies, expression profiling and other low-scale experimental studies. To evaluate the evidence, we collected metadata about each study including clinical and demographic features, experimental design and statistical significance, and used a scoring and ranking approach to select a core data set of 434 high-confidence genes. The genes mapped to pathways including neuroactive ligand–receptor interaction, synapse transmission and axon guidance. To better understand the genes we parsed over 30 databases to retrieve extensive data about expression patterns, protein interactions, animal models and pharmacogenetics. We constructed a MySQL-based online database and share it with the broader autism research community at http://autismkb.cbi.pku.edu.cn, supporting sophisticated browsing and searching functionalities.
INTRODUCTION
Autism spectrum disorder (ASD) is a heterogeneous neurodevelopmental disorder characterized by impairments in reciprocal social interaction and communication and presence of restricted, repetitive and stereotyped patterns of behavior, interests and activities (1). ASD is an umbrella term for Autistic Disorder, Asperger Syndrome and Pervasive Developmental Disorder Not Otherwise Specified (PDD-NOS) (1). With an early onset prior to age 3 and a prevalence as high as 0.9–2.6% (2,3), ASD is one of the leading causes of childhood disability and inflicts serious suffering and burden for the family and society (4).
Understanding the causes of ASD is critical for developing better treatment. Twin studies have shown that the heritability of ASD is as high as 38–90%, indicating strong contributions by genetic factors as well as environmental factors (5,6). The search for environmental factors has not yet led to convincing major candidates whereas the search for genes associated with autism, although far from complete or conclusive, has been more fruitful. The genes discovered so far can be roughly grouped into two categories: ‘syndromic autism related genes’ or causal genes underlying genetic disorders that cause autistic symptoms such as Fragile X Syndrome, Rett Syndrome, Tuberous Sclerosis Complex and dozens of other disorders (7,8), and ‘non-syndromic autism related genes’ most of which are susceptibility genes (9). Many experimental methods have been used to identify associated genes, including the earlier linkage analyses and low-scale candidate gene association or experimental studies as well as the more recent genome-wide association studies (GWAS), genome-wide CNV studies and expression profiling.
With hundreds of studies published, especially the recent genome-wide studies, and with the next-generation sequencing technologies providing even more power for further gene discoveries (10), a new challenge has emerged: it has become more and more difficult for an autism researcher to answer with confidence how many genes have been associated with ASD, how strong the evidence is, what features the genes have and what pathways they involve. The amount of available literature and data and the intrinsic complexity of autism genetics demand bioinformatic data management and analysis. Three efforts have been made so far by different groups to collect genes and variations associated with ASD: AutDB (also known as SAFRI Gene) collected 219 genes (11,12), Autism genetic database (AGD) collected 226 genes and 743 CNVs (13) and Autism Chromosome Rearrangement Database (ACRD) collected 372 breakpoints and other genomic features (14). However, they are far from a comprehensive survey of autism genetics. To bring a clearer big picture of autism genetics, we performed a comprehensive review and analysis of published literature and data, described below, resulting in a total of 2193 genes, 2806 SNPs/VNTRs, 4544 CNVs and 158 linkage regions. We provide the results as an online resource for the broader autism research community at http://autismkb.cbi.pku.edu.cn/ with extensive evidence and annotations, supporting sophisticated browsing and searching functionalities.
DATA COLLECTION
Literature search
We searched the PubMed database for publications related to autism genetics, using the query term ‘autism AND associat*’ for association studies, ‘autism AND (gene OR microarray OR proteomics)’ for expression profiling studies and the other low-scale experimental studies, and ‘autism AND (CNV OR copy number variation OR microarray* OR microdel* OR microdup* OR rearrange* OR (genome-wide AND (linkage OR associa* OR scan)))’ for CNV and linkage studies. The abstracts of the 4000+ articles retrieved were reviewed to remove irrelevant papers, resulting in a final set of 579 articles, reporting a total of 11 GWAS, 242 low-scale candidate gene association studies, 13 expression profiling studies, 95 genome-wide CNV studies, 23 genome-wide linkage analyses and 236 other low-scale experimental studies.
For syndromic autism-related genes, we first collected the autism-related disorders and their causal genes from a recently published comprehensive review (7). We then searched OMIM to get the official disease names and linked all the disorders to OMIM, and searched PubMed for additional citations using the query ‘(OMIM disease name) AND autism’ for each disease. All citations were double-checked manually. Finally, 99 genes for 94 autism-related disorders supported by 250 references were included in our data set of ‘Syndromic Autism Related Genes’.
In total, we collected as many as 2135 non-syndromic autism-related genes, 99 syndromic autism-related genes, 4544 CNVs and 158 linkage regions. The genes located in the CNV and linkage regions were then retrieved by the UCSC Genome Browser (15).
Evidence collection
To establish the strength of evidence, we collected metadata about each study and result. Supplementary Table S1–S7 list the evidence collected for each type of experimental methods. In summary, for each study of non-syndromic autism, we collected the clinical and demographic features of the samples including ancestral background, country of origin, inclusion and exclusion criteria, number of cases and controls with gender ratio, age at examination and diagnosis criteria. We collected metadata about the experimental design including platform, experimental methods, statistical methods and statistical significance.
For each gene, we estimated how much evidence supports its role in autism by each type of experimental methods and calculated a weighted sum, following a multi-dimensional evidence-based candidate gene prioritization approach (16). First, we assigned initial scores to the genes for each type of experimental methods (Supplementary Table S8). Score 0 is given if there is no positive evidence of the type. Table 1 lists the distribution of the scores for each type. Next, we used a benchmark data set consisting of 21 non-syndromic autism-related genes considered high confidence from six autism reviews (8,9,17–20) (Supplementary Table S9) to calculate the weights. We followed a gene prioritization approach (16) to generate a candidate weight matrix pool consisting of dN = 76 weight vectors, where N represents the number of experimental methods and d = N+1 represents possible different weights, 1–7 in the weight vectors. A combined score for each gene was then calculated by summing up the products of the scores and corresponding weights from the six experimental methods (16). All the 2135 candidate genes including 21 benchmark genes were sorted by their combined scores. We selected the weight matrix that gave the benchmark genes the highest rank as the optimal weight matrix (Supplementary Table S10). About 95% benchmark genes were ranked among the top 98% of all candidate genes. We chose the lowest combined score, 9, from the benchmark data set as the cutoff of high-confident genes, resulting in a core data set of 383 non-syndromic autism-related genes. Because the definition of ‘optimal weight matrix’ is always debatable, we provide an online ranking tool to allow users to re-rank the genes interactively by inputting customized weights based on their own experiences and preferences.
Table 1.
Score distribution of genes discovered by each experiment method
| Experimental methods | Scores | Number of Genes |
|---|---|---|
| Genome-wide association studies | 1 | 81 |
| 2 | 46 | |
| 3 | 5 | |
| Expression profiling | 1 | 1320 |
| 2 | 285 | |
| 3 | 50 | |
| Genome-wide CNV studies | 1 | 1086 |
| 2 | 34 | |
| 3 | 19 | |
| Linkage analyses | 1 | 535 |
| 2 | 43 | |
| 3 | 0 | |
| Low scale genetic association studies | 1 | 128 |
| 2 | 23 | |
| 3 | 12 | |
| Other low-scale experimental studies | 1 | 241 |
| 2 | 37 | |
| 3 | 30 |
For syndromic autism, we assigned four levels to the autism-related disorders: Level 1 disorders have one reported case with autistic symptoms, Level 2 have two to three cases in a single family, Level 3 have cases in more than one family and Level 4 are reported in multiple review papers (8). Causal genes of Level 3 and 4 disorders were considered high-confident genes in the core dataset.
Functional annotations
To better understand the function of the genes associated with autism, we collected extensive functional information and data, including crosslinks to NCBI Entrez gene (21), OMIM (21), Uniprot (http://www.uniprot.org/) and Ensembl (http://www.ensembl.org/), functional groups based on Gene Ontology (http://www.geneontology.org/), protein–protein interactions from database BioGRID (22), BIND (23) and HPRD (24), and genomic variants from the Database of Genomic Variants (DGV) (25). We linked the genes to three psychiatric disease databases, AlzGene (26), SzGene (27) and PDGene (http://www.pdgene.org/), when the gene is common between these diseases and ASD. Information about homologues of the genes were retrieved from Mouse Genome Informatics (MGI) (28), Zebrafish Model Organism Database (ZFIN) (29) and FlyBase (30). We collected comprehensive mRNA expression profiling data, including ESTs from NCBI Unigene Profiles (21), microarray expression profiles from BioGPS (31) and Allen Brain Atlas (32), and RNA-Seq (33–38). Protein expression evidence at peptide level was retrieved from PRIDE (39) and Peptide Atlas (40). We also collected transcription factor binding sites in the upstream regions of the genes from in-house collection of ChIP-Chip and ChIP-Seq data, miRNAs that may target the genes from miRWalk (41) and TarBase (42), and natural antisense transcripts that may regulate the genes from NATsDB (43). Possible post-translation modifications were retrieved from UniProt and dbPTM (44). We used KOBAS 2.0 (45) to retrieve the pathways that the genes are involved in from BioCyc (46), KEGG Pathway (47), PID (48), PID Reactome (48), PANTHER (49) and Reactome (50) and possible association with other diseases from Disease databases include KEGG Disease (51), FunDO (52,53), GAD (54), NHGRI GWAS Catalog (55) and OMIM (21). Pharmaco-genetics and drug information was collected from Comparative Toxicogenomics Database (CTD) (56), Pharmacogenomics Knowledge Base (57) and DrugBank (58). Supplementary Table S11 summarizes the gene coverage from each source database. The overlap between the genes discovered by expression profiling and those by the other genetic technologies is shown in Supplementary Table S12.
Enriched functional pathways were identified by KOBAS 2.0 (45) and enriched GO terms were identified by DAVID (59). Pathways such as neuroactive ligand–receptor interaction, synapse transmission, and axon guidance were statistically significantly enriched in the core data set (Table 2). In addition to synapse transmission, GO terms such as transmission of nerve impulse, neuron differentiation were also found to be statistically significant (Table 3). The result is consistent with recent findings that synapse development, axon targeting and neuron motility are related to autism etiology (60,61).
Table 2.
Top five enriched pathway of the genes in the high-confident core dataset, using KOBAS2.0
| Term | Database | ID | P Value | Q Value |
|---|---|---|---|---|
| Neuroactive ligand-receptor interaction | KEGG PATHWAY | hsa04080 | 1.03E-11 | 1.65E-09 |
| Synaptic Transmission | Reactome | REACT:13685 | 7.50E-10 | 9.06E-08 |
| Axon guidance | Reactome | REACT:18266 | 1.29E-08 | 1.24E-06 |
| Calcium signaling pathway | KEGG PATHWAY | hsa04020 | 2.25E-08 | 1.97E-06 |
| Long-term potentiation | KEGG PATHWAY | hsa04720 | 1.76E-07 | 9.98E-06 |
Table 3.
Top 10 enriched GO terms of the genes in the high-confident core dataset
| GO ID | GO Term | P Value | Q Value |
|---|---|---|---|
| GO:0019226 | transmission of nerve impulse | 5.44E-29 | 9.73E-26 |
| GO:0007268 | synaptic transmission | 4.59E-28 | 8.21E-25 |
| GO:0007610 | synapse | 1.05E-23 | 1.45E-20 |
| GO:0045202 | behavior | 4.53E-23 | 8.10E-20 |
| GO:0044057 | synapse part | 7.21E-22 | 9.94E-19 |
| GO:0007267 | regulation of system process | 4.12E-21 | 7.38E-18 |
| GO:0044456 | cell-cell signaling | 4.17E-21 | 7.46E-18 |
| GO:0030182 | neuron differentiation | 8.21E-19 | 1.47E-15 |
| GO:0031644 | regulation of neurological system process | 1.53E-18 | 2.74E-15 |
| GO:0051969 | regulation of transmission of nerve impulse | 1.74E-18 | 3.11E-15 |
DATABASE INTERFACE
We set up a MySQL relational database to store all the data. A user-friendly web interface for browsing and searching was implemented by PHP and JavaScript, powered by JQuery framework.
Browsing
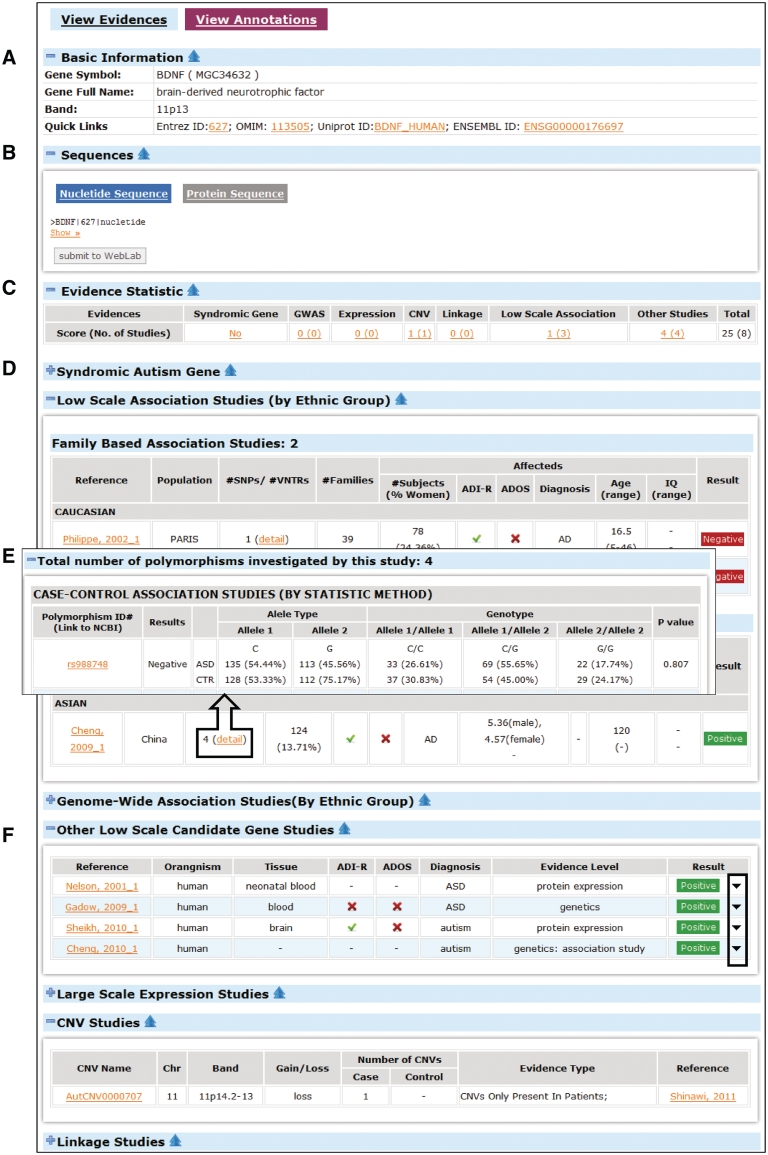
Users can browse the data in AutismKB in a variety of ways, including by data sets, experimental methods or chromosome. The gene lists include a summary of information about the genes, hyperlinked to detailed gene evidence and annotation pages. Figure 1 shows a typical AutismKB gene entry. Basic information such as gene symbol, gene name, cytoband and cross links are provided (Figure 1A). Nucleotide sequences and protein sequences can be sent to WebLab (62) for further analysis (Figure 1B). Summaries of supporting evidence and category-specific scores are provided (Figure 1C). Users can click on the hyperlinks of the category-specific score to view different category of evidences. The categories without any evidence are hidden by default (Figure 1D). Users can click on ‘+’ to expand or ‘−’ to collapse different categories. Detailed information of polymorphisms for low scale association studies and GWAS can be found by clicking on ‘detail’ in the tables (Figure 1E). When exploring other low-scale studies and large-scale expression studies, users can click the down arrow in the right of the table to obtain more information (Figure 1F). Annotations of each gene can be obtained by clicking the label ‘view annotation’ in the top left.
Figure 1.
A typical gene entry in AutismKB. (A) Basic information and quick links, (B) nucleotide and protein sequences, (C) evidence statistics and links to different data sources, (D) example of default collapsed data source, (E) link and example of polymorphism information and (F) example of expanded data source with hidden information.
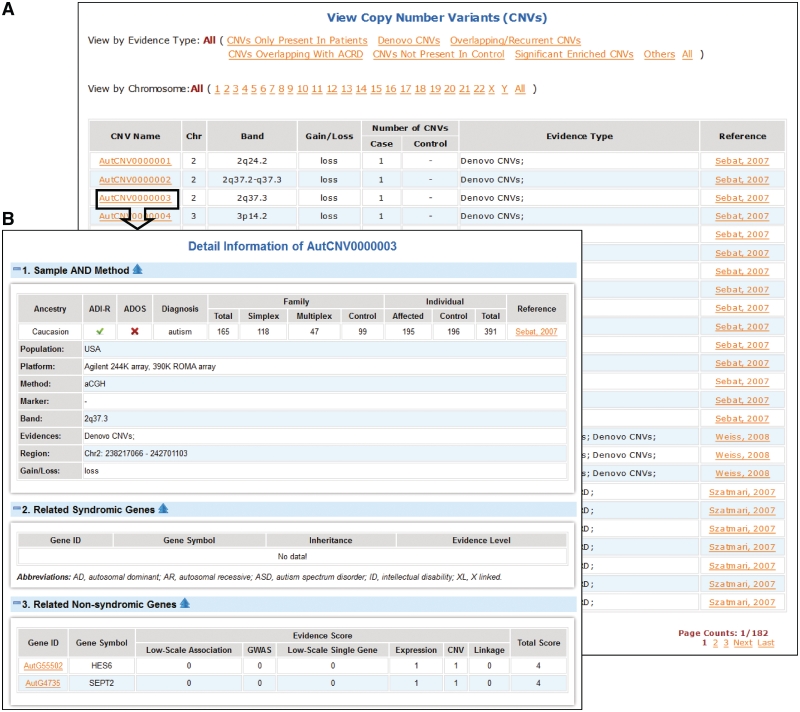
CNVs are provided by a tabular view with name, cytoband, gain or loss, number, evidence types and reference. Users can use evidence type and chromosome to filter the table (Figure 2A). Clicking on the name can bring the detail information of each CNV including the samples and methods of the study, CNV region, and any syndromic and non-syndromic autism genes in the region (Figure 2B). Users can use chromosome to filter the linkage regions and click on linkage name to view detailed information.
Figure 2.
CNV list and a typical CNV entry in AutismKB. (A) the CNV list in AutismKB and (B) a typical CNV entry.
Searching
AutismKB supports both text-based search and sequence-based search. Users can find a quick search box on the top right of each page to search by gene symbol. Advanced search was provided to search genes, CNVs, linkage regions by gene name, gene symbol, NCBI Entrez id, Ensemble id, GO terms, UniProt ID, location, score, method and PubMed ID. Finally, a BLAST search against the nucleotide or protein sequences of all AutismKB genes is also available.
CONCLUSION
AutismKB is a comprehensive knowledgebase of autism-related genes, CNVs and linkage regions with extensive evidence and annotations. AutismKB will be updated periodically. We hope that it can be a valuable resource for the autism research community.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1-12.
FUNDING
Funding for open access charge: National Outstanding Young Investigator Award from Natural Science Foundation of China (grant number: 31025014); 973 Basic Research Program (grant number: 2011CBA01102); scholarships from Merck and Johnson and Johnson.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank Ge Gao, Chuan-Yun Li, Yong-Xin Ye and Ying-Fu Zhong for useful comments on the web interface.
REFERENCES
- 1.American Psychiatric Association. Diagnostic and Statistical Manual of Mental Disorders: DSM-IV-TR. Arlington, VA: American Psychiatric Publishing, Inc; 2000. [Google Scholar]
- 2.Kogan MD, Blumberg SJ, Schieve LA, Boyle CA, Perrin JM, Ghandour RM, Singh GK, Strickland BB, Trevathan E, van Dyck PC. prevalence of parent-reported diagnosis of autism spectrum disorder among children in the US, 2007. Pediatrics. 2009;124:1395–1403. doi: 10.1542/peds.2009-1522. [DOI] [PubMed] [Google Scholar]
- 3.Kim YS, Leventhal BL, Koh YJ, Fombonne E, Laska E, Lim EC, Cheon KA, Kim SJ, Kim YK, Lee H, et al. Prevalence of autism spectrum disorders in a total population sample. Am. J. Psychiatry. 2011;168:904–912. doi: 10.1176/appi.ajp.2011.10101532. [DOI] [PubMed] [Google Scholar]
- 4.Ganz ML. The Costs of Autism. In: Moldin SO, Rubenstein JLR, editors. Understanding Autism: from Basic Neuroscience to Treatment. Boca Raton, FL: CRC Press; 2006. pp. 476–498. [Google Scholar]
- 5.Hallmayer J, Cleveland S, Torres A, Phillips J, Cohen B, Torigoe T, Miller J, Fedele A, Collins J, Smith K, et al. Genetic heritability and shared environmental factors among twin pairs with autism. Arch. Gen. Psychiatry. 2011;68:1095–1102. doi: 10.1001/archgenpsychiatry.2011.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bailey A, Le Couteur A, Gottesman I, Bolton P, Simonoff E, Yuzda E, Rutter M. Autism as a strongly genetic disorder: evidence from a British twin study. Psychol. Med. 1995;25:63–77. doi: 10.1017/s0033291700028099. [DOI] [PubMed] [Google Scholar]
- 7.Betancur C. Etiological heterogeneity in autism spectrum disorders: more than 100 genetic and genomic disorders and still counting. Brain Res. 2011;1380:42–77. doi: 10.1016/j.brainres.2010.11.078. [DOI] [PubMed] [Google Scholar]
- 8.Abrahams BS, Geschwind DH. Advances in autism genetics: on the threshold of a new neurobiology. Nat. Rev. Genet. 2008;9:341–355. doi: 10.1038/nrg2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.State MW. The genetics of child psychiatric disorders: focus on autism and Tourette syndrome. Neuron. 2010;68:254–269. doi: 10.1016/j.neuron.2010.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lander ES. Initial impact of the sequencing of the human genome. Nature. 2011;470:187–197. doi: 10.1038/nature09792. [DOI] [PubMed] [Google Scholar]
- 11.Banerjee-Basu S, Packer A. SFARI Gene: an evolving database for the autism research community. Dis. Model Mech. 2010;3:133–135. doi: 10.1242/dmm.005439. [DOI] [PubMed] [Google Scholar]
- 12.Basu SN, Kollu R, Banerjee-Basu S. AutDB: a gene reference resource for autism research. Nucleic Acids Res. 2009;37:D832–D836. doi: 10.1093/nar/gkn835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Matuszek G, Talebizadeh Z. Autism Genetic Database (AGD): a comprehensive database including autism susceptibility gene-CNVs integrated with known noncoding RNAs and fragile sites. BMC Med. Genet. 2009;10:102. doi: 10.1186/1471-2350-10-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marshall CR, Noor A, Vincent JB, Lionel AC, Feuk L, Skaug J, Shago M, Moessner R, Pinto D, Ren Y, et al. Structural variation of chromosomes in autism spectrum disorder. Am. J. Hum. Genet. 2008;82:477–488. doi: 10.1016/j.ajhg.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fujita PA, Rhead B, Zweig AS, Hinrichs AS, Karolchik D, Cline MS, Goldman M, Barber GP, Clawson H, Coelho A, et al. The UCSC Genome Browser database: update 2011. Nucleic Acids Res. 2011;39:D876–D882. doi: 10.1093/nar/gkq963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sun J, Jia P, Fanous AH, Webb BT, van den Oord EJ, Chen X, Bukszar J, Kendler KS, Zhao Z. A multi-dimensional evidence-based candidate gene prioritization approach for complex diseases-schizophrenia as a case. Bioinformatics. 2009;25:2595–6602. doi: 10.1093/bioinformatics/btp428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Freitag CM. The genetics of autistic disorders and its clinical relevance: a review of the literature. Mol. Psychiatry. 2007;12:2–22. doi: 10.1038/sj.mp.4001896. [DOI] [PubMed] [Google Scholar]
- 18.Klauck SM. Genetics of autism spectrum disorder. Eur. J. Hum. Genet. 2006;14:714–720. doi: 10.1038/sj.ejhg.5201610. [DOI] [PubMed] [Google Scholar]
- 19.Losh M, Sullivan PF, Trembath D, Piven J. Current developments in the genetics of autism: from phenome to genome. J. Neuropathol. Exp. Neurol. 2008;67:829–837. doi: 10.1097/NEN.0b013e318184482d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Muhle R, Trentacoste SV, Rapin I. The genetics of autism. Pediatrics. 2004;113:e472–e486. doi: 10.1542/peds.113.5.e472. [DOI] [PubMed] [Google Scholar]
- 21.Sayers EW, Barrett T, Benson DA, Bolton E, Bryant SH, Canese K, Chetvernin V, Church DM, DiCuccio M, Federhen S, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2011;39:D38–51. doi: 10.1093/nar/gkq1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stark C, Breitkreutz BJ, Chatr-Aryamontri A, Boucher L, Oughtred R, Livstone MS, Nixon J, Van Auken K, Wang X, Shi X, et al. The BioGRID Interaction Database: 2011 update. Nucleic Acids Res. 2011;39:D698–704. doi: 10.1093/nar/gkq1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gilbert D. Biomolecular interaction network database. Brief Bioinform. 2005;6:194–198. doi: 10.1093/bib/6.2.194. [DOI] [PubMed] [Google Scholar]
- 24.Keshava Prasad TS, Goel R, Kandasamy K, Keerthikumar S, Kumar S, Mathivanan S, Telikicherla D, Raju R, Shafreen B, Venugopal A, et al. Human Protein Reference Database—2009 update. Nucleic Acids Res. 2009;37:D767–D772. doi: 10.1093/nar/gkn892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang J, Feuk L, Duggan GE, Khaja R, Scherer SW. Development of bioinformatics resources for display and analysis of copy number and other structural variants in the human genome. Cytogenet Genome Res. 2006;115:205–214. doi: 10.1159/000095916. [DOI] [PubMed] [Google Scholar]
- 26.Bertram L, McQueen MB, Mullin K, Blacker D, Tanzi RE. Systematic meta-analyses of Alzheimer disease genetic association studies: the AlzGene database. Nat. Genet. 2007;39:17–23. doi: 10.1038/ng1934. [DOI] [PubMed] [Google Scholar]
- 27.Allen NC, Bagade S, McQueen MB, Ioannidis JP, Kavvoura FK, Khoury MJ, Tanzi RE, Bertram L. Systematic meta-analyses and field synopsis of genetic association studies in schizophrenia: the SzGene database. Nat. Genet. 2008;40:827–834. doi: 10.1038/ng.171. [DOI] [PubMed] [Google Scholar]
- 28.Blake JA, Bult CJ, Kadin JA, Richardson JE, Eppig JT. The Mouse Genome Database (MGD): premier model organism resource for mammalian genomics and genetics. Nucleic Acids Res. 2011;39:D842–D848. doi: 10.1093/nar/gkq1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bradford Y, Conlin T, Dunn N, Fashena D, Frazer K, Howe DG, Knight J, Mani P, Martin R, Moxon SA, et al. ZFIN: enhancements and updates to the Zebrafish Model Organism Database. Nucleic Acids Res. 2011;39:D822–D829. doi: 10.1093/nar/gkq1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tweedie S, Ashburner M, Falls K, Leyland P, McQuilton P, Marygold S, Millburn G, Osumi-Sutherland D, Schroeder A, Seal R, et al. FlyBase: enhancing Drosophila Gene Ontology annotations. Nucleic Acids Res. 2009;37:D555–D559. doi: 10.1093/nar/gkn788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Su AI, Wiltshire T, Batalov S, Lapp H, Ching KA, Block D, Zhang J, Soden R, Hayakawa M, Kreiman G, et al. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc. Natl Acad. Sci. USA. 2004;101:6062–6067. doi: 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jones AR, Overly CC, Sunkin SM. The Allen Brain Atlas: 5 years and beyond. Nat. Rev. Neurosci. 2009;10:821–828. doi: 10.1038/nrn2722. [DOI] [PubMed] [Google Scholar]
- 33.Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu JQ, Habegger L, Noisa P, Szekely A, Qiu C, Hutchison S, Raha D, Egholm M, Lin H, Weissman S, et al. Dynamic transcriptomes during neural differentiation of human embryonic stem cells revealed by short, long, and paired-end sequencing. Proc. Natl Acad. Sci. USA. 2010;107:5254–5259. doi: 10.1073/pnas.0914114107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 37.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vizcaino JA, Cote R, Reisinger F, Foster JM, Mueller M, Rameseder J, Hermjakob H, Martens L. A guide to the Proteomics Identifications Database proteomics data repository. Proteomics. 2009;9:4276–4283. doi: 10.1002/pmic.200900402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Farrah T, Deutsch EW, Omenn GS, Campbell DS, Sun Z, Bletz JA, Mallick P, Katz JE, Malmstrom J, Ossola R, et al. A high-confidence human plasma proteome reference set with estimated concentrations in PeptideAtlas. Mol. Cell Proteomics. 2011;10:M110 006353. doi: 10.1074/mcp.M110.006353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dweep H, Sticht C, Pandey P, Gretz N. miRWalk - Database: Prediction of possible miRNA binding sites by ‘walking’ the genes of three genomes. J. Biomed Inform. 2011;44:839–847. doi: 10.1016/j.jbi.2011.05.002. [DOI] [PubMed] [Google Scholar]
- 42.Papadopoulos GL, Reczko M, Simossis VA, Sethupathy P, Hatzigeorgiou AG. The database of experimentally supported targets: a functional update of TarBase. Nucleic Acids Res. 2009;37:D155–D158. doi: 10.1093/nar/gkn809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang Y, Liu XS, Liu QR, Wei L. Genome-wide in silico identification and analysis of cis natural antisense transcripts (cis-NATs) in ten species. Nucleic Acids Res. 2006;34:3465–3475. doi: 10.1093/nar/gkl473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee TY, Hsu JB, Chang WC, Wang TY, Hsu PC, Huang HD. A comprehensive resource for integrating and displaying protein post-translational modifications. BMC Res. Notes. 2009;2:111. doi: 10.1186/1756-0500-2-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xie C, Mao X, Huang J, Ding Y, Wu J, Dong S, Kong L, Gao G, Li CY, Wei L. KOBAS 2.0: a web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 2011;39:W316–W322. doi: 10.1093/nar/gkr483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Karp PD, Ouzounis CA, Moore-Kochlacs C, Goldovsky L, Kaipa P, Ahren D, Tsoka S, Darzentas N, Kunin V, Lopez-Bigas N. Expansion of the BioCyc collection of pathway/genome databases to 160 genomes. Nucleic Acids Res. 2005;33:6083–6089. doi: 10.1093/nar/gki892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kanehisa M, Araki M, Goto S, Hattori M, Hirakawa M, Itoh M, Katayama T, Kawashima S, Okuda S, Tokimatsu T, et al. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008;36:D480–D484. doi: 10.1093/nar/gkm882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schaefer CF, Anthony K, Krupa S, Buchoff J, Day M, Hannay T, Buetow KH. PID: the Pathway Interaction Database. Nucleic Acids Res. 2009;37:D674–D679. doi: 10.1093/nar/gkn653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thomas PD, Campbell MJ, Kejariwal A, Mi H, Karlak B, Daverman R, Diemer K, Muruganujan A, Narechania A. PANTHER: a library of protein families and subfamilies indexed by function. Genome Res. 2003;13:2129–2141. doi: 10.1101/gr.772403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Croft D, O'Kelly G, Wu G, Haw R, Gillespie M, Matthews L, Caudy M, Garapati P, Gopinath G, Jassal B, et al. Reactome: a database of reactions, pathways and biological processes. Nucleic Acids Res. 2011;39:D691–D697. doi: 10.1093/nar/gkq1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kanehisa M, Goto S, Furumichi M, Tanabe M, Hirakawa M. KEGG for representation and analysis of molecular networks involving diseases and drugs. Nucleic Acids Res. 2010;38:D355–D360. doi: 10.1093/nar/gkp896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Osborne JD, Flatow J, Holko M, Lin SM, Kibbe WA, Zhu LJ, Danila MI, Feng G, Chisholm RL. Annotating the human genome with Disease Ontology. BMC Genomics. 2009;10(Suppl. 1):S6. doi: 10.1186/1471-2164-10-S1-S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Du P, Feng G, Flatow J, Song J, Holko M, Kibbe WA, Lin SM. From disease ontology to disease-ontology lite: statistical methods to adapt a general-purpose ontology for the test of gene-ontology associations. Bioinformatics. 2009;25:i63–i68. doi: 10.1093/bioinformatics/btp193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Becker KG, Barnes KC, Bright TJ, Wang SA. The genetic association database. Nat. Genet. 2004;36:431–432. doi: 10.1038/ng0504-431. [DOI] [PubMed] [Google Scholar]
- 55.Hindorff LA, Sethupathy P, Junkins HA, Ramos EM, Mehta JP, Collins FS, Manolio TA. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc. Natl Acad. Sci USA. 2009;106:9362–9367. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Davis AP, King BL, Mockus S, Murphy CG, Saraceni-Richards C, Rosenstein M, Wiegers T, Mattingly CJ. The Comparative Toxicogenomics Database: update 2011. Nucleic Acids Res. 2011;39:D1067–D1072. doi: 10.1093/nar/gkq813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Relling MV, Gardner EE, Sandborn WJ, Schmiegelow K, Pui CH, Yee SW, Stein CM, Carrillo M, Evans WE, Klein TE. Clinical Pharmacogenetics Implementation Consortium guidelines for thiopurine methyltransferase genotype and thiopurine dosing. Clin. Pharmacol. Ther. 2011;89:387–391. doi: 10.1038/clpt.2010.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Knox C, Law V, Jewison T, Liu P, Ly S, Frolkis A, Pon A, Banco K, Mak C, Neveu V, et al. DrugBank 3.0: a comprehensive resource for ‘omics’ research on drugs. Nucleic Acids Res. 2011;39:D1035–D1041. doi: 10.1093/nar/gkq1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 60.Gilman SR, Iossifov I, Levy D, Ronemus M, Wigler M, Vitkup D. Rare de novo variants associated with autism implicate a large functional network of genes involved in formation and function of synapses. Neuron. 2011;70:898–907. doi: 10.1016/j.neuron.2011.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Voineagu I, Wang X, Johnston P, Lowe JK, Tian Y, Horvath S, Mill J, Cantor RM, Blencowe BJ, Geschwind DH. Transcriptomic analysis of autistic brain reveals convergent molecular pathology. Nature. 2011;474:380–384. doi: 10.1038/nature10110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Liu X, Wu J, Wang J, Zhao S, Li Z, Kong L, Gu X, Luo J, Gao G. WebLab: a data-centric, knowledge-sharing bioinformatic platform. Nucleic Acids Res. 2009;37:W33–W39. doi: 10.1093/nar/gkp428. [DOI] [PMC free article] [PubMed] [Google Scholar]