Abstract
UniPathway (http://www.unipathway.org) is a fully manually curated resource for the representation and annotation of metabolic pathways. UniPathway provides explicit representations of enzyme-catalyzed and spontaneous chemical reactions, as well as a hierarchical representation of metabolic pathways. This hierarchy uses linear subpathways as the basic building block for the assembly of larger and more complex pathways, including species-specific pathway variants. All of the pathway data in UniPathway has been extensively cross-linked to existing pathway resources such as KEGG and MetaCyc, as well as sequence resources such as the UniProt KnowledgeBase (UniProtKB), for which UniPathway provides a controlled vocabulary for pathway annotation. We introduce here the basic concepts underlying the UniPathway resource, with the aim of allowing users to fully exploit the information provided by UniPathway.
INTRODUCTION
Dealing with the metabolic network of a living organism as a whole is extremely complex, and so it is commonly broken down into smaller parts or subnetworks, called metabolic pathways. Pathways are often defined or thought of as the elementary functional and evolutionary building blocks of the complete metabolic network, with each pathway being a ‘self-contained’ elementary biochemical process. However, no universal and clear-cut definition of metabolic pathways exists. Any attempt to partition the reaction network of an organism into a set of (possibly overlapping) metabolic pathways will require some arbitrary decisions as to where such partitions should be made or how pathway variants should be described. As pointed out by Green and Karp (1), the same network can be described using different rationalizations (or conceptualizations) of pathways, each of which meets a specific user need. It is therefore important to explicitly describe the concepts that are used in the construction of a particular pathway database to allow the user to fully understand and exploit the resource. In the following section, we highlight the major features of some existing pathway-related resources, namely KEGG (2), MetaCyc (3) and the SEED (4–6). These features are illustrated by a comparison of how each of these resources represents the variant pathways that result in the biosynthesis of l-lysine. We then introduce the major conceptual features of the UniPathway resource and illustrate how UniPathway is used for pathway annotation of individual proteins in UniProtKB.
Representation of the l-lysine biosynthesis pathway in existing pathway resources
l-lysine can be produced de novo in prokaryotes, lower eukaryotes and some plants by two distinct biosynthetic pathways [see (7) for recent review]: the meso-diaminopimelate (DAP) pathway (in archaea, bacteria, lower fungi and plants), and the l-α-aminoadipate (AAA) pathway (in archaea, deinococci, dictyostelium and higher fungi). Four different variants of the DAP pathway have been identified [see (8) for review]. All DAP variants have l-aspartate as precursor and share the initial and terminal steps but differ in the production of the ll-2,6-diaminopimelate and dl-2,6-diaminopimelate intermediates. Two different variants of the AAA pathway also give rise to l-lysine from 2-oxoglutarate (α-ketoglutarate) via l-α-aminoadipate (9,10). Hence, four DAP variant pathways and two AAA variant pathways are known to give rise to l-lysine. We now describe how this variant pathway information is represented in the KEGG, MetaCyc and SEED resources, and contrast these representations of l-lysine biosynthesis with that of UniPathway.
KEGG (2) provides chemical information (compounds and reactions), genomic information (genes, genomes, species and groups of orthologs) and pathways. In KEGG, the metabolic pathways—called ‘maps’—are subparts of the overall reaction graph. Reactions within a map are connected by their constituent metabolites, which also provide links to reactions in other maps. KEGG metabolic maps are described without reference to a particular species, and each map includes the reactions belonging to all known variants of a particular pathway. KEGG represents the process of l-lysine biosynthesis using a single map (http://www.genome.jp/dbget-bin/www_bget?pathway:map00300), including all biochemical reactions relating to l-lysine biosynthesis and without distinguishing between the DAP and AAA pathways and their variants. Supplementary Figure S1 shows the corresponding KEGG map in which individual subpathways within the map have been highlighted. The color scheme used is identical to that used in Figure 1, where each color corresponds to one distinct ‘linear subpathway’ (described in detail below) in the process of l-lysine biosynthesis. The use of a common color scheme is intended to facilitate the identification of common subpathways within the different representations of l-lysine biosynthesis that are provided by the various resources.
Figure 1.
Representation of the l-lysine biosynthesis pathway in UniPathway. The l-lysine biosynthesis pathway is specialized in two chemically defined pathway variants (the DAP and AAA pathways) by an ‘IsA’ relationship. The DAP pathway is composed of seven linear subpathways (ULS) and the AAA pathway is composed of three linear subpathways (which are ‘PartOf' their respective pathway variants). Colored boxes, using the same color code as in the Supplementary figures for comparisons, indicate the subpathways. The right part of the figure presents an exploded view of the first linear subpathway (ULS) of the AAA pathway, which is composed of four Enzymatic reactions (UERs).
MetaCyc is a database of non-redundant, experimentally elucidated metabolic pathways from many species (3). The related resource EcoCyc provides similar information for Escherichia coli, and was historically one of the first attempts to conceptualize metabolic data in a rigorous way (11). MetaCyc explicitly represents and stores pathways as well as compounds, proteins, protein complexes and genes. MetaCyc specifically defines individual pathway variants and assigns unique identifiers to them. MetaCyc represents the ‘Lysine biosynthesis’ pathway as six different ‘Lysine biosynthesis’ variants, termed I, II, III, IV, V and VI. Of these 6 variants, I, II, III and VI are specific to the DAP pathway, while variants IV and V are specific to the AAA pathway. In MetaCyc, these pathways are independent but can be related through their common metabolites and reactions. Supplementary Figure S2 shows the MetaCyc pathway variants for l-lysine biosynthesis, with subpathways highlighted according to the common coloring scheme.
The SEED (4–6) is a comparative genomics environment primarily devoted to the annotation of genomic data and the construction of genome-scale metabolic models. Annotation is performed using expert-curated ‘subsystems’, where each subsystem is defined as a set of ‘functional roles’ that make up a biological process such as a metabolic pathway, and where the scope or limits of the subsystem in question are defined by the curator (4). The SEED describes l-lysine biosynthesis in two distinct and independent subsystems, one for the DAP pathway and one for the AAA pathway (see Supplementary Figure S3). This representation lies somewhere between that of KEGG (one single map), and MetaCyc (six different variants). Subsystems can be further divided into reaction subnetworks or ‘scenarios’, where each scenario represents a set of connected reactions that convert a defined set of substrates into a defined set of products (4). Scenarios may include additional reactions outside those of the subsystem in which the scenario occurs (such as spontaneous reactions), and can be used to identify points that connect individual subsystems during the process of metabolic network reconstruction by the Model SEED pipeline (5,6). The DAP pathway subsystem is further subdivided into two consecutive scenarios describing the conversion of l-aspartate to meso-2,6-diaminoheptanedioate and the subsequent conversion of meso-2,6-diaminoheptanedioate to l-lysine.
Representation of the l-lysine biosynthesis pathway in UniPathway
The pathway concepts described above correspond to different and complementary viewpoints on metabolism. While KEGG may provide metabolic maps including all known pathway variants, the SEED may break these down into distinct subsystems and scenarios, and MetaCyc into individual pathway variants. UniPathway adopts concepts from resources like KEGG, MetaCyc and SEED, including the idea of pathway variants, but incorporates additional concepts designed to make the description of species-specific pathway variants more applicable to protein annotation. A full description of the concepts underlying the UniPathway resource is provided in the following section, but we would here like to draw the attention of the reader to one key concept, that of the UniPathway Linear Subpathway, or ULS. Each ULS represents a linear succession of enzymatic reactions that are known to be connected as a series, and for which no variant is currently known. The ULS can therefore be considered as the basic building block for the assembly of larger pathways and pathway variants. By breaking down large pathways into their constituent units, UniPathway avoids the requirement to specifically instantiate each pathway variant as a separate entity. Instead, pathway variants are represented as alternate paths through a set of connected ULS. Each ULS is named using its endpoint compounds, producing a controlled vocabulary for use in enzyme annotation. To illustrate how a pathway is constructed from combinations of individual ULS, we consider again the process of l-lysine biosynthesis. This process is described within UniPathway as two different metabolic pathways: the DAP pathway and the AAA pathway (Figure 1), which are both specializations of the ‘l-lysine biosynthesis’ term. The DAP variant pathways and the AAA variant pathways are themselves composed of specific combinations of linear subpathways (ULS), with seven distinct ULS contributing to the four DAP variant pathways and three distinct ULS contributing to the two AAA variant pathways, as illustrated in Figure 1.
UNIPATHWAY CONCEPTS
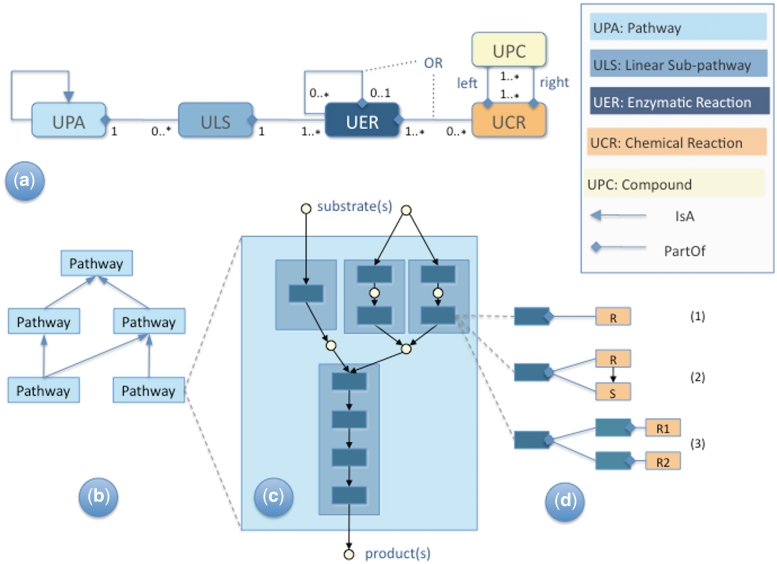
In this section, we present a more detailed description of the concepts underlying the UniPathway resource. Following the guidelines given by Green and Karp (1), we use the term ‘pathway conceptualization’ to denote the explicit description of pathways as physical processes composed of chemical reactions and compounds. This requires definition of the reaction components, the relationships between them, and the start and end point of each pathway. An overview of the UniPathway conceptualization is given in Table 1 and in Figure 2a in the form of a simplified Unified Modeling Language (UML) diagram (12). The major entities of UniPathway are the ‘Compound (UPC)’, ‘Chemical Reaction (UCR)’, ‘Enzymatic Reaction (UER)’, ‘Linear Subpathway (ULS)’ and ‘Pathway (UPA)’.
Table 1.
UniPathway classes and their attributes
| UniPathway classes | Mandatory attributes | Optional attributes |
|---|---|---|
| UPC compound |
|
|
| UCR chemical reaction |
|
|
| UER enzymatic reaction |
|
|
| ULS linear subpathway |
|
|
| UPA pathway |
|
|
Figure 2.
Overview of the UniPathway concepts. (a) Unified Modeling Language (UML)-like representation of the UniPathway classes and relationships. Legend is to the right of the main part of the figure. Multiplicity constraints read as: One UPA is composed of 0 or more ULS—One ULS is contained in exactly 1 UPA. One ULS is composed of 1 or more UER—One UER is contained in exactly 1 ULS. One UER is composed of 0 or more (alternate) UER—One UER is contained in 0 or at most 1 UER. One UER is composed of 0 or more UCR—One UCR is contained in 1 or more UER. One UCR is composed of 1 or more left UPC and 1 or more right UPC—One UPC is contained in 1 or more UCR. (b) Example of the IsA relationship defining the UniPathway controlled vocabulary hierarchy of pathway terms. A pathway instance may be a specific type of an abstract pathway entity. (c) Example of the PartOf relationship linking a pathway (UPA: light blue), its subpathways (ULS: blue) and individual enzymatic reactions that constitute the subpathway (UER: dark blue). (d) Three cases of the relationship between an UER and its chemical reaction components (UCR): (1) simple one-to-one relationship where R is catalyzed by a single enzyme; (2) R is catalyzed by an enzyme and S is a spontaneous reaction; (3) ‘OR’ relationship: the enzyme can catalyze two reactions differing by their co-substrates (e.g. NADH/NADPH).
A ‘Compound (UPC)’ is the lowest level chemical entity involved in a biochemical reaction. It can be a low molecular-weight molecule, a polymer or a biopolymer (a protein or a nucleic acid). Some compounds may correspond to abstract entities such as an alcohol, or DNA.
A ‘Chemical Reaction (UCR)’ is an irreducible chemical transformation of a multi-set of chemical compounds to another multi-set of chemical compounds. ‘Irreducible’ means that the reaction cannot be split into smaller subreactions (as far as chemical knowledge permits). ‘Multi-set’ simply means that we keep track of the number of times each compound appears on each side of the reaction (i.e. its stoichiometry). In UniPathway, a UCR is always considered as reversible. Therefore, the choice of which compounds are represented on the left or right side of the reaction is arbitrary. This definition is strictly identical to the one used in KEGG.
An ‘Enzymatic Reaction (UER)’ is a chemical transformation catalyzed by an enzyme. In UniPathway, the UER is a central concept since it represents transformations that are directly linked to (and referenced by) proteins, defined by UniProtKB entries (13). UERs are directly associated to UniProtKB entries (not indirectly linked through EC numbers), although a UER may be linked to an EC number. Distinct UERs may include the same reaction (UCR), if that reaction happens to be catalyzed by different enzymes. UERs are only defined within the context of a given linear subpathway (i.e. ULS), which allows us to name them in a rational way (see below). Most UERs are associated to a single UCR [Figures 2d(1) and 3a] and correspond to a single catalytic reaction (with a single EC number). UERs can also be associated with UCR(s) corresponding to spontaneous reactions, providing such reactions immediately follow (or precede) the catalyzed reaction [Figures 2d(2) and 3b]. When a catalytic reaction actually corresponds to two (or more) alternate reactions, all of them being catalyzed by the same enzyme [Figure 2d(3)], we represent this as two different alternate reactions (as in KEGG). In practice, this is implemented by a set of alternate UERs, each of which is associated either to a single or multiple UCRs in order to represent any combinatorial composition. Such cases can occur when the enzyme uses alternate co-substrates or co-products (such as NADH or NADPH) while the ‘main’ substrates and products remain the same. This contrasts with NC-IUBMB and MetaCyc which describe one enzyme class and one reaction with an abstract compound [such as NAD(P)H].
Figure 3.
Example of relationships between ULS, UER and UCR. ULS00012—‘l-α-aminoadipate from 2-oxoglutarate’—is a linear subpathway composed of four UERs linked through their primary compounds. (a) The first step in ULS00012 is UER00028, associated to the chemical reaction UCR00271 (using Left-to-Right direction). This UCR involves five compounds, but only two of these, 2-oxoglutarate and (R)-homocitrate, are considered to be primary compounds in the context of UER00028. (b) The second step in ULS00012 is UER00029, associated to two chemical reactions: UCR03444 (using Left-to-Right direction) followed by UCR04371 (using Right-to-Left direction). The primary substrate of UER00029 is (R)-homocitrate and its primary product is homoisocitrate.
A ‘Linear Subpathway (ULS)’, also simply called ‘subpathway’, is a chemical transformation from a multi-set of initial compounds [substrate(s)] to a multi-set of final compounds [product(s)] that does not contain any branching reaction or cycle (Figure 2c). More precisely, if we define the ‘reaction graph’ as a graph where vertices are reactions and two vertices are linked by an edge where the product of one reaction is the substrate of the next, then ULS are simply paths in this graph. Technically, an ULS is therefore an ordered sequence of UERs. UERs within a ULS are linked via their primary metabolites, which are defined according to the context of the pathway. For example, in Figure 3, for the linear subpathway ULS00012, ‘l-α-aminoadipate from 2-oxoglutarate’, the primary product of the reaction UER00028 is (R)-homocitrate. This product links UER00028 to the following reaction UER00029, of which it is the primary substrate. In UniPathway, we have defined each ULS according to the principle of parsimony, that is, we have defined the smallest set of ULS that allows the decomposition of all known pathways. This means that as more reactions and pathways are added to UniPathway the existing ULS definitions will evolve to accommodate this new information. For example, the discovery of a new variant in an existing ULS would mean that this ULS would have to be split accordingly. For reaction cycles, we decided to split them into two (or more) ULS at arbitrarily selected points. Individual UERs within an ULS are assigned a ‘step number’ from 1 to n, where n is the total number of reactions in the ULS.
A ‘Pathway (UPA)’ is generally composed of a set of linear subpathways (ULS), connected through their common compounds. Each ULS is found in only one pathway, which facilitates protein annotation using ULS and their parent pathway terms (see also the section ‘UniPathway as a tool for annotation of UniProtKB/Swiss-Prot’). The set of ULS for a given pathway can be empty; allowing the definition of abstract pathways (such as the ‘amino-acid biosynthesis pathway’) as well as pathways whose precise composition is as yet unknown. In this way, UniPathway provides a hierarchical terminology of pathways (Figure 2b) similar to the Gene Ontology (14) ‘biological process’ namespace (to which it has been mapped), and, like GO terms, UniPathway terms have been defined to facilitate cross-species annotation. We saw how higher order pathway definitions were used to group the two sets of l-lysine biosynthesis pathways, ‘l-lysine biosynthesis’ via ‘DAP pathway’, and ‘l-lysine biosynthesis’ via ‘AAA pathway’, which are both concrete instances of ‘l-lysine biosynthesis’ and ‘amino-acid biosynthesis’ pathways (Figure 1).
UNIPATHWAY IMPLEMENTATION AND DATA SOURCES
The UniPathway schema is implemented within the PostgreSQL (8.2) relational DBMS (http://www.postgresql.org/). The UniPathway database is populated with primary chemical data (UPC, UCR) that is imported from KEGG LIGAND (2). Enzymatic reactions, subpathways and pathways (UER, ULS, UPA) are manually curated by UniPathway curators. This curation process makes use of primary literature and data from existing metabolic resources such as KEGG and MetaCyc. Curation involves: checking reaction stoichiometry, linking reactions to the appropriate UniProtKB entries, defining the start and end points and constituent reactions of linear subpathways (ULS), assembling ULS into pathways (UPA) (which requires definition of pathway endpoints and topology), and the curation of pathway names. Finally, direct links from UER, ULS and UPA to external resources are also manually created. This currently involves adding PubMed identifiers for bibliography, Gene Ontology (GO) terms and MetaCyc pathways. The links to KEGG maps are computed automatically based on the UCR-KEGG reaction cross-references. UniPathway is also linked indirectly via the UniProtKB associations provided by each UER to a host of additional resources including InterPro (15), Prosite (16), HAMAP (17), Pfam (18) and PRIAM (19), as well as Genome Reviews genes (20) and the NCBI taxonomy (21). Table 2 summarizes the current content of UniPathway (release 2011_08 of July 2011).
Table 2.
UniPathway content (release 2011_08 of July 2011)
| UniPathway classes | Number of instances |
|---|---|
| UPA: pathway | 1007 (including 270 pathways defined at the level of reactions) |
| ULS: linear subpathway | 493 |
| UER: enzymatic reaction | 1009 |
| UCR: chemical reaction | 986 |
| UPC: compound | 1087 |
UNIPATHWAY WEB SITE AND DISTRIBUTION
UniPathway is accessible through a dedicated web server at the following URL: http://www.unipathway.org
The portal allows users to search UniPathway data using simple textual terms as well as identifiers including EC numbers, UniProtKB accession numbers, or GO terms. It provides a number of specific views for each data type, including:
a ‘chemical perspective’, displaying the chemical structure of the object (e.g. a reaction graph);
a ‘protein perspective’, exploiting the UniProtKB/Swiss-Prot entries associated to specific reactions, and providing information such as the distribution of protein/domain families, UniProtKB keywords, GO terms, etc;
a ‘genomic perspective’, displaying, for a chosen species, the genomic context of the genes involved in a pathway; and
a ‘taxonomic perspective’, summarizing in the form of a table or tree, the presence/absence of reactions, subpathways or pathways within selected species or other taxonomic groups.
UniPathway data is also distributed as flat files in OBO 1.2 format (http://www.geneontology.org/GO.format.obo-1_2.shtml) or as tabulated files at http://www.unipathway.org/download/unipathway. This data is updated and synchronized at each UniProtKB release.
UNIPATHWAY AS A TOOL FOR ANNOTATION OF UNIPROTKB/SWISS-PROT
UniPathway provides a structured controlled vocabulary for pathways that uses universal, linear subpathways as the basic building block for higher order pathway assemblies. These linear subpathways can be used to annotate individual proteins in the absence of a complete genome sequence. This makes UniPathway eminently suitable for the annotation of pathway information within UniProtKB protein records, many of which are not associated with a complete genome sequence.
Within UniProtKB, pathway information is provided in the ‘Pathways’ subsection of the ‘General annotation’ section in the following form (as viewed in flat text):
CC -!- PATHWAY: SuperPathway; Pathway(; SubPathway: EnzReaction) ([regulation]).
Where ( ) indicates optional fields.
The ‘EnzReaction’ field describes the enzymatic reaction (UER) to which this entry is actually linked to, while the ‘SubPathway’ field describes the linear subpathway (ULS) of which the UER is a part. The ‘Pathway’ field describes the pathway (UPA) of which the ULS is in turn a part, and the ‘SuperPathway’ field is an abstract parent term of Pathway in the UniPathway hierarchical controlled vocabulary (such as ‘amino-acid biosynthesis’). Terms for ‘SuperPathway’ and ‘Pathway’ are defined by curators within the UniPathway controlled vocabulary, where the ‘SuperPathway’ term chosen for annotation can be any one of the parent terms lying between the Pathway and the root. This ‘SuperPathway’ term must be sufficiently general to allow cross-species annotation of all UniProtKB proteins within a particular UER (and the ULS and UPA of which it is a part). Terms for ‘SubPathway’ are automatically created from the list of initial substrate(s) and final product(s) of the ULS using the following syntax:
product (and product)+ from substrate (and substrate)+
where ‘substrate’ and ‘product’ are the labels (common name) of the corresponding compound (UPC) in UniPathway. Since each ULS is a linear sequence of UERs, the ‘EnzReaction’ field is simply written as the step number of the particular UER in that ULS, according to the format: ‘step n/m’, where ‘n’ is the step number and ‘m’ the total number of steps in the ULS. Note that both the ‘SubPathway’ and ‘EnzReaction’ fields are optional, and may be absent where detailed biochemical reaction(s) are not yet known or curated. Finally, the ‘regulation’ keyword indicates that the protein acts as a transcriptional regulator of the genes coding for enzymes of the pathway, but this information is still scarce in the current version of the database.
The following are typical examples of CC-PATHWAY records that appear in UniProtKB/Swiss-Prot entries of the release current at time of writing (release 2011_08):
P49367
CC -!- PATHWAY: Amino-acid biosynthesis; L-lysine biosynthesis via AAA
CC pathway; L-alpha-aminoadipate from 2-oxoglutarate: step 2/4.
P0A877
CC -!- PATHWAY: Amino-acid biosynthesis; L-tryptophan biosynthesis; L-
CC tryptophan from chorismate: step 5/5.
P95477
CC -!- PATHWAY: Siderophore biosynthesis; pseudomonine biosynthesis.
P52957
CC -!- PATHWAY: Mycotoxin biosynthesis; sterigmatocystin biosynthesis
CC [regulation].
UniProtKB/Swiss-Prot records P49367 and P0A877 contain complete pathway annotations including ‘SuperPathway’, ‘Pathway’, ‘SubPathway’ and step number. For both these records, the chosen ‘SuperPathway’ term is the general term for amino acid biosynthesis, rather than the direct parent of the named Pathway (which is ‘l-lysine biosynthesis’). UniProtKB/Swiss-Prot record P95477 corresponds to a partially characterized activity, where the enzyme is known to be involved in pseudomonine (siderophore) biosynthesis, but where detailed information on the chemical reaction is not available. Finally, the UniProtKB/Swiss-Prot record P52957 describes a transcriptional regulator of sterigmatocystin biosynthesis.
UniPathway has been used to provide a controlled vocabulary for pathway annotation within UniProtKB records from UniProt release 14.7 (January 2009). Metabolic pathway information flows from UniPathway to UniProt. UniProt curators use the existing UniPathway controlled vocabulary to annotate proteins and can, when necessary, request new pathway definitions from UniPathway curators. UniPathway data is then used as a reference to control further metabolic pathway annotations in UniProt. In release 2011_08 of UniProtKB, UniPathway provided annotation for 118 390 distinct UniProtKB/Swiss-Prot protein records and 783 299 UniProtKB/TrEMBL protein records. Each of these UniProtKB records is linked, via the ‘Pathway’ subsection of the ‘General annotation’ section, to the appropriate pathway description within the UniPathway web site.
CONCLUSION AND FUTURE DIRECTIONS
UniPathway is a resource for the representation and annotation of enzymatic reactions and metabolic pathways. UniPathway provides an explicit biochemical description of each reaction, allowing individual reactions to be linked via their chemical constituents, and reduces each metabolic pathway to a set of constituent linear subpathways, or ULS. Sets of interlinked ULS are then assembled into a larger pathway, or UPA, which can in turn be assembled into larger pathways. UniPathway avoids the need to enumerate individual pathway variants while providing a hierarchical controlled vocabulary for pathways that allows related pathway assemblies to be easily recognized. UniPathway provides pathway annotation for UniProtKB protein records, where a specific combination of reaction (UER), linear subpathway (ULS) and pathway (UPA) define the role of a protein. UniPathway thereby provides a direct link from proteins (enzymes) in UniProtKB to known biochemical reactions, without the need to link them indirectly through EC numbers. UniPathway also serves as a stand-alone reference resource on metabolism for a number of projects relating to metabolic network reconstruction such as the Microme (http://www.microme.eu) and MetaNetX (http://www.metanetx.org) initiatives.
We will continue to maintain and improve the UniPathway resource and the underlying data model. Planned improvements include the addition of curated information on protein complexes and subcellular locations, which may be necessary for the correct definition of enzyme requirements and compartmentalized pathways. One limitation with the current model is encountered when defining pathways that have a large number of alternative routes (such as the pathways leading to the production of secondary metabolites in plants). Such pathways will be reduced to a large number of short ULS, and in extreme cases, ULS composed of a single reaction (UER). While in such cases the notion of a ULS may be less useful, well defined alternative routes could still be described by connecting these ULS/UER into a pathway (UPA) and assigning a specific name to that pathway.
In the near future, UniPathway will switch to using ChEBI (22) as the primary source for chemical data and Rhea (http://www.ebi.ac.uk/rhea/) as the primary source of reaction data (Rhea itself being based on ChEBI), although links to other metabolic resources will continue to be provided. This change will improve consistency between chemical structures and labels and will allow users access to the underlying chemical ontology of ChEBI, but may affect compound labeling (as Rhea and KEGG represent chemical entities at different pH values).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online. Supplementary Figures 1–3.
FUNDING
Swiss Federal Government through the Federal Office of Education and Science, European Union (SLING: Serving Life-science Information for the Next Generation: 226073, Microme: A Knowledge-Based Bioinformatics Framework for Microbial Pathway Genomics: 222886-2 and ERC Advanced Grant SISYPHE), French government through ANR MIRI BLAN08-1335497 and MetaNetX project of the Swiss SystemsX.ch initiative. Computational hardware resources were provided by the Pôle Rhône-Alpin de Bioinformatique and funded by the GIS-IBISA. IT support was provided by INRIA-Rhône-Alpes. Funding for open access charge: Swiss Federal Government through the Federal Office of Education and Science.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We would like to thank Guillaume Lelaurain from INRIA IT support staff for his continuous commitment and, Frédéric Boyer, Sophie Huet and Adrien Maudet for their invaluable help in the early stages of this project. We gratefully thank Professor Minoru Kanehisa for permission to use his data. We would also like to thank the reviewers for their helpful comments and suggestions for improvements to the manuscript.
REFERENCES
- 1.Green ML, Karp PD. The outcomes of pathway database computations depend on pathway ontology. Nucleic Acids Res. 2006;34:3687–3697. doi: 10.1093/nar/gkl438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kanehisa M, Goto S, Furumichi M, Tanabe M, Hirakawa M. KEGG for representation and analysis of molecular networks involving diseases and drugs. Nucleic Acids Res. 2010;38:D355–D360. doi: 10.1093/nar/gkp896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Caspi R, Altman T, Dale JM, Dreher K, Fulcher CA, Gilham F, Kaipa P, Karthikeyan AS, Kothari A, Krummenacker M, et al. The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases. Nucleic Acids Res. 2010;38:D473–D479. doi: 10.1093/nar/gkp875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Overbeek R, Begley T, Butler RM, Choudhuri JV, Chuang HY, Cohoon M, de Crécy-Lagard V, Diaz N, Disz T, Edwards R, et al. The subsystems approach to genome annotation and its use in the project to annotate 1000 genomes. Nucleic Acids Res. 2005;33:5691–5702. doi: 10.1093/nar/gki866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.DeJongh M, Formsma K, Boillot P, Gould J, Rycenga M, Best A. Toward the automated generation of genome-scale metabolic networks in the SEED. BMC Bioinformatics. 2007;8:139. doi: 10.1186/1471-2105-8-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Henry CS, DeJongh M, Best AA, Frybarger PM, Linsay B, Stevens RL. High throughput generation, optimization and analysis of genome-scale metabolic models. Nat. Biotech. 2010;9:977–982. doi: 10.1038/nbt.1672. [DOI] [PubMed] [Google Scholar]
- 7.Dairi T, Kuzuyama T, Nishiyama M, Fujii I. Convergent strategies in biosynthesis. Nat. Prod. Rep. 2011;28:1054–1086. doi: 10.1039/c0np00047g. [DOI] [PubMed] [Google Scholar]
- 8.Hudson AO, Gilvarg C, Leustek T. Biochemical and phylogenetic characterization of a novel diaminopimelate biosynthesis pathway in prokaryotes identifies a diverged form of LL-diaminopimelate aminotransferase. J. Bacteriol. 2008;190:3256–3263. doi: 10.1128/JB.01381-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kosuge T, Hoshino T. Lysine is synthesized through the alpha-aminoadipate pathway in Thermus thermophilus. FEMS Microbiol. Lett. 1998;169:361–367. doi: 10.1111/j.1574-6968.1998.tb13341.x. [DOI] [PubMed] [Google Scholar]
- 10.Horie A, Tomita T, Saiki A, Kono H, Taka H, Mineki R, Fujimura T, Nishiyama C, Kuzuyama T, Nishiyama M. Discovery of proteinaceous N-modification in lysine biosynthesis of Thermus thermophilus. Nat. Chem. Biol. 2009;5:673–679. doi: 10.1038/nchembio.198. [DOI] [PubMed] [Google Scholar]
- 11.Karp PD, Riley M, Paley SM, Pelligrini-Toole A. EcoCyc: an encyclopedia of Escherichia coli genes and metabolis. Nucleic Acids Res. 1996;24:32–39. doi: 10.1093/nar/24.1.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Webb K, White T. UML as a cell and biochemistry modeling language. Biosystems. 2005;80:283–302. doi: 10.1016/j.biosystems.2004.12.003. [DOI] [PubMed] [Google Scholar]
- 13.The UniProt Consortium. Ongoing and future developments at the Universal Protein Resource. Nucleic Acids Res. 2011;39:D214–D219. doi: 10.1093/nar/gkq1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gene Ontology Consortium. The Gene Ontology in 2010: extensions and refinements. Nucleic Acids Res. 2010;38:D331–D335. doi: 10.1093/nar/gkp1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hunter S, Apweiler R, Attwood TK, Bairoch A, Bateman A, Binns D, Bork P, Das U, Daugherty L, et al. InterPro: the integrative protein signature database. Nucleic Acids Res. 2009;37:D224–D228. doi: 10.1093/nar/gkn785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sigrist CJ, Cerutti L, de Castro E, Langendijk-Genevaux PS, Bulliard V, Bairoch A, Hulo N. PROSITE, a protein domain database for functional characterization and annotation. Nucleic Acids Res. 2010;38:D161–D166. doi: 10.1093/nar/gkp885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lima T, Auchincloss AH, Coudert E, Keller G, Michoud K, Rivoire C, Bulliard V, de Castro E, Lachaize C, Baratin D, et al. HAMAP: a database of completely sequenced microbial proteome sets and manually curated microbial protein families in UniProtKB/Swiss-Prot. Nucleic Acids Res. 2009;37:D471–D478. doi: 10.1093/nar/gkn661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Finn RD, Mistry J, Tate J, Coggill P, Heger A, Pollington JE, Gavin OL, Gunesekaran P, Ceric G, Forslund K, et al. The Pfam protein families database. Nucleic Acids Res. 2010;38:D211–D222. doi: 10.1093/nar/gkp985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Claudel-Renard C, Chevalet C, Faraut T, Kahn D. Enzyme-specific profiles for genome annotation: PRIAM. Nucleic Acids Res. 2003;31:6633–6639. doi: 10.1093/nar/gkg847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kersey P, Bower L, Morris L, Horne A, Petryszak R, Kanz C, Kanapin A, Das U, Michoud K, Phan I, et al. Integr8 and genome reviews: integrated views of complete genomes and proteomes. Nucleic Acids Res. 2005;33:D297–D302. doi: 10.1093/nar/gki039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sayers EW, Barrett T, Benson DA, Bryant SH, Canese K, Chetvernin V, Church DM, DiCuccio M, Edgar R, Federhen S, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2009;37:D5–D15. doi: 10.1093/nar/gkn741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.de Matos P, Alcántara R, Dekker A, Ennis M, Hastings J, Haug K, Spiteri I, Turner S, Steinbeck C. Chemical entities of biological interest: an update. Nucleic Acids Res. 2010;38:D249–D254. doi: 10.1093/nar/gkp886. [DOI] [PMC free article] [PubMed] [Google Scholar]