Abstract
Accurate structure-based sequence alignments of distantly related proteins are crucial in gaining insight about protein domains that belong to a superfamily. The PASS2 database provides alignments of proteins related at the superfamily level and are characterized by low sequence identity. We thus report an automated, updated version of the superfamily alignment database known as PASS2.4, consisting of 1961 superfamilies and 10 569 protein domains, which is in direct correspondence with SCOP (1.75) database. Database organization, improved methods for efficient structure-based sequence alignments and the analysis of extreme distantly related proteins within superfamilies formed the focus of this update. Alignment of family-specific functional residues can be realized using such alignments and is shown using one superfamily as an example. The database of alignments and other related features can be accessed at http://caps.ncbs.res.in/pass2/.
INTRODUCTION
The motivation for improved protein structure comparison, alignment and characterization is currently defined simply by quantity-the rate of increase in the number of experimentally determined new folds and the number of structures adopting each fold. Accurate sequence alignments for homologous proteins are essential for constructing accurate motifs, profiles and in building homology models (1). The correct sequence alignment of distantly related proteins, where the sequence similarity is very low, is often hard to obtain based on sequence similarity alone (2,3). In such cases, structure-based sequence alignment methods could be helpful to reveal features that are essential for both structure and function. The observation of structural homology leads to the development of structural alignment tools, which are becoming useful upon the acceleration of protein structure determination and the Structural Genomics project (4).
Protein domains that are grouped together at superfamily level are defined as having structural, functional and sequence similarities and evidence for a common evolutionary ancestor. They are also characterized by conserved structural core and poor sequence identity. SCOP (5) database provides a detailed and comprehensive description about protein structures organized at different hierarchies of structural and functional similarities. ASTRAL (6) provides an explicit mapping between the PDB ATOM and SEQRES records within PDB files, which is used to derive databases of sequences corresponding to the SCOP domains. A somewhat similar database as ours is S4 (7), which provides multiple structure-based alignments of SCOP (version 1.63) protein superfamilies and was made publicly available in the year 2005. There are well known databases available for alignment of homologous proteins. The HOMSTRAD (8) database contains aligned three-dimensional structures of homologous proteins. PALI (9) is another database providing Phylogeny and ALIgnment of homologous protein structures and contains structure-based sequence alignments. PASS2 database provides structure-based sequence alignments of the SCOP superfamilies and it is updated according to the SCOP release since 1998. Here, we report an updated version of the PASS2 version 4 in direct correspondence with the SCOP 1.75. Besides a simple update with accumulated entries (as described in ‘Overview of PASS2 versions’ below), we have modified the codes to handle large superfamilies. The codes have now been organized in Linux platform for convenient updates in future and our alignment protocol employs improved methods of alignment. We have explained about the mapping of family-specific functional residues using riboflavin synthase superfamily as an example. We have also analysed the extreme-deviant members, the outliers, of some superfamilies.
OVERVIEW OF PASS2 VERSIONS
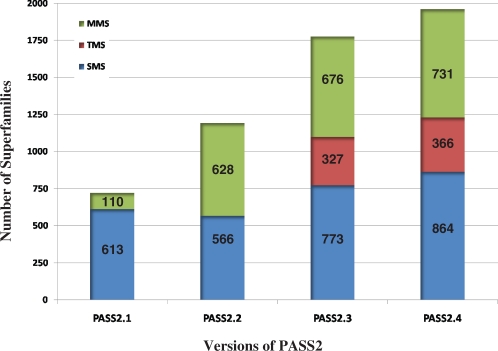
The idea of structure-based sequence alignment and analysis of protein domain superfamilies originally started with CAMPASS (10), The automated version of CAMPASS, called as PASS2 (11), which we now refer to as PASS2.1, contained 613 superfamilies in direct correspondence with SCOP 1.53. The subsequent versions of PASS2 [PASS2.2 and PASS2.3 (12,13)] have been updated in direct correspondence with SCOP1.63 and SCOP 1.73, respectively. In most PASS2 versions, we have classified the superfamilies into single-member (SMS), two-member (TMS) and multi-member (MMS) superfamilies, which directly implies the number of domains with <40% identity with other domains in the superfamily. TMS and MMS are aligned using specific alignment method from PASS2 version 3 onwards. The statistics of all the four versions are reported in Figure 1. The current version of PASS2, PASS2.4, holds 10 569 protein domains (at a 40% sequence identity cut-off) belonging to 1961 superfamilies and is in direct correspondence with SCOP 1.75.
Figure 1.
Overview of PASS2 over the past few versions. Number of superfamilies from PASS2.1 (8), PASS2.2 (9) through PASS2.3 (10) have increased over the years. Total number of superfamilies are shown in SMS, TMS and MMS categories.
IMPROVEMENTS IN THE CURRENT VERSION
PASS2 version 4 is updated in correspondence with the SCOP 1.75. Alignment protocol has been revised as described in alignment protocol. This version of database also aims at improved user interface, like JMOL view, JMOL command input area and introductory pop-ups for search results. In continuation of our introduction of the outliers in PASS2.3, in the current version, we have re-examined the nature and category of outliers in superfamilies (Supplementary Data). In the earlier versions, there were difficulties in aligning large superfamilies. These issues have been addressed so that it is possible to automate the whole protocol and move the codes to the Linux platform. The protocol is being automated for further updates to minimize any manual interventions.
ALIGNMENT PROTOCOL
Initially, pre-processing of the domains such as removing the hetero atoms and retaining one coordinate set in NMR structure are done using in-house programmes. For TMS, MINRMS (14) is used for the initial alignment and that initial equivalences are utilized by COMPARER (15) for the refined alignment. After a careful assessment of different protocols for the alignment of MMS (detailed in Supplementary Data, Supplementary Tables ST1, ST2 and Supplementary Figure SF1), MATT (16) was chosen for initial alignment. From the initial alignment, equivalent regions were identified by JOY (17) and structure-guided tree information was obtained from MATT to form as inputs in COMPARER. These initial equivalences serve as seeds for rigid-body superposition using MNFC, a modified form of MNYFIT (18) (Supplementary Figure SF2). Final accepted alignments were structure annotated for the structural information such as, secondary structural regions, solvent accessibility of residues and pattern of hydrogen bonds by employing the JOY program. The alignment is assessed using mean RMSD and percentage of conserved secondary structural equivalence (POCSSE) (Supplementary Data).These two parameters were viewed as important quality checks of multiple alignment.
ORGANIZATION OF THE DATABASE
Similar to the previous versions (Supplementary Table ST3), the major focus of database is at the superfamily level, but searches can be made using keywords at various levels, like SCOP classes, folds and domains. The current version, PASS2.4, provides information about features such as HMM (19,20), Structural Motif (21), structural phylogeny, PCA analysis and CUSP (22,23) as discussed in the previous versions of PASS2.2 and PASS2.3. In addition, all the feature files, alignments and structural superposition are downloadable via webpage. At the protein domain level, accessory files, used for JOY (17), like PSA, SST and HBD files are also downloadable. Other utilities such as, PSI-BLAST (24), PHI-BLAST (24), constructed HMM profiles based on PASS2 alignments and 3D structural annotation of query alignment/sequence, were modified and updated corresponding to the latest PASS2 database. Some general utilities such as Alistat (19), multiple formats of the alignment and a README file, which is helpful for the user to know more details about the each superfamily are also provided as in the previous version.
MAPPING FAMILY-SPECIFIC FUNCTIONAL RESIDUE MOTIFS: EXAMPLE OF RIBOFLAVIN SYNTHASE SUPERFAMILY
Protein function prediction is one of the central problems in computational biology. The confidence of function inference from structure depends on the number of family-specific motifs found in the query structure compared to their distribution in a large non-redundant database of proteins (25). The PASS2 protocol is able to map the family specific as well as functional important residues. We have done the case study on riboflavin synthase superfamily which consists of three families. After a careful structure-based alignment of superfamily members, as recorded in PASS2.4, the motif pattern of LTV and VNV are specific to only riboflavin synthase family (26,27) and pattern GD and GQ are specific to NADPH-cytochrome p450 reductase FAD-binding domain-like family and reductase FAD-binding domain-like family, respectively.
The results show that our structure-based sequence alignment protocol retains family specific as well as functionally important residues in equivalent positions in the alignment. This is one of the important applications of the PASS2 alignments that show the critical analysis of superfamilies and functionally important as well as family-specific residues is possible (riboflavin synthase superfamily in Supplementary Figure SF3).
CONCLUSIONS
PASS2 database organizes structure-based sequence alignments of protein domain superfamilies in correspondence with SCOP definitions. In this update of PASS2 database, PASS2.4, we have introduced maximal level of automation. In addition, PASS2.4 alignments were useful to align functionally important residues as well as family-specific residues (Supplementary Figure SF4–SF6). We also suggest that structurally deviant superfamily members could be removed as outliers, so that such extreme distant relationships will not influence the alignment. Analysis of structural and sequence differences amongst known superfamily members hopefully provide useful
guidelines for modelling distantly related proteins.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–3, Supplementary Figures 1–6 and Supplementary References [28,29].
FUNDING
Funding for open access charge: Department of Biotechnology, India.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
A.G is supported by Senior Research Fellowship from the Council of Scientific and Industrial Research (CSIR) Government of India. We thank Dr Kanagarajadurai and Ms. S. Kalaimathy for helpful discussions. We thank Department of Biotechnology, India and NCBS for financial and infrastructural support.
REFERENCES
- 1.Hubbard TJP, Blundell TL. Comparison of solvent-inaccessible cores of homologous proteins: definitions useful for protein modelling. Protein Eng. 1987;1:159–71. doi: 10.1093/protein/1.3.159. [DOI] [PubMed] [Google Scholar]
- 2.Sauder JM, Arthur JW, Dunbrack RL., Jr Large-scale comparison of protein sequence alignment algorithms with structure alignments. Proteins. 2000;40:6–22. doi: 10.1002/(sici)1097-0134(20000701)40:1<6::aid-prot30>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 3.Marchler-Bauer A, Panchenko AR, Ariel N, Bryant SH. Comparison of sequence and structure alignments of protein domains. Proteins. 2002;48:439–46. doi: 10.1002/prot.10163. [DOI] [PubMed] [Google Scholar]
- 4.Koehl P. Protein structure similarities. Curr. Opin. Struct. Biol. 2001;11:348–353. doi: 10.1016/s0959-440x(00)00214-1. [DOI] [PubMed] [Google Scholar]
- 5.Andreeva A, Howorth D, Chandonia JM, Brenner SE, Hubbard TJ, Chothia C, Murzin AG. Data growth and its impact on the SCOP database: new developments. Nucleic Acids Res. 2008;36:D419–425. doi: 10.1093/nar/gkm993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chandonia JM, Hon G, Walker NS, Lo Conte L, Koehl P, Levitt M, Brenner SE. The ASTRAL compendium in 2004. Nucleic Acids Res. 2004;32:D189–D192. doi: 10.1093/nar/gkh034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Casbon J, Saqi MA. S4: structure-based sequence alignments of SCOP superfamilies. Nucleic Acids Res. 2005;33:D219–D222. doi: 10.1093/nar/gki043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stebbings LA, Mizuguchi K. HOMSTRAD: recent developments of the Homologous Protein Structure Alignment Database. Nucleic Acids Res. 2004;32:D203–D207. doi: 10.1093/nar/gkh027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Balaji S, Sujatha S, Sai Chetan Kumar S, Srinivasan N. PALI - A database of Phylogeny and ALIgnment of homologous protein structures. Nucleic Acids Res. 2001;29:61–65. doi: 10.1093/nar/29.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sowdhamini R, Burke DF, Huang JF, Mizuguchi K, Nagarajaram HA, Srinivasan N, Steward RE, Blundell TL. CAMPASS: a database of structurally aligned protein superfamilies. Structure. 1998;6:1087–1094. doi: 10.1016/s0969-2126(98)00110-5. [DOI] [PubMed] [Google Scholar]
- 11.Mallika V, Bhaduri A, Sowdhamini R. PASS2: a semi-automated database of protein alignments organized as structural superfamilies. Nucleic Acids Res. 2002;30:284–288. doi: 10.1093/nar/30.1.284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bhaduri A, Pugalenthi G, Sowdhamini R. PASS2: an automated database of protein alignments organized as structural superfamilies. BMC Bioinformatics. 2004;5:35. doi: 10.1186/1471-2105-5-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kanagarajadurai K, Kalaimathy S, Nagarajan P, Sowdhamini R. PASS2, a database of structure-based sequence alignments of protein structural domain sperfamilies: towards automatic updation. IJKDB. 2011 (in press) [Google Scholar]
- 14.Jewett AI, Huang CC, Ferrin TE. MINRMS: an efficient algorithm for determining protein structure similarity using root-mean-squared-distance. Bioinformatics. 2003;19:625–634. doi: 10.1093/bioinformatics/btg035. [DOI] [PubMed] [Google Scholar]
- 15.Sali A, Blundell TL. Definition of general topological equivalence in protein structures. A procedure involving comparison of properties and relationships through simulated annealing and dynamic programming. J. Mol. Biol. 1990;212:403–428. doi: 10.1016/0022-2836(90)90134-8. [DOI] [PubMed] [Google Scholar]
- 16.Menke M, Berger B, Cowen L. Matt: local flexibility aids protein multiple structure alignment. PloS Comput. Biol. 2008;4:e10. doi: 10.1371/journal.pcbi.0040010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mizuguchi K, Deane CM, Blundell TL, Johnson MS, Overington JP. JOY: protein sequence-structure representation and analysis. Bioinformatics. 1998;14:617–623. doi: 10.1093/bioinformatics/14.7.617. [DOI] [PubMed] [Google Scholar]
- 18.Sutcliffe MJ, Haneef I, Carney D, Blundell TL. Knowledge based modelling of homologous proteins, Part I: Three-dimensional frameworks derived from the simultaneous superposition of multiple structures. Protein Eng. 1987;1:377–384. doi: 10.1093/protein/1.5.377. [DOI] [PubMed] [Google Scholar]
- 19.Eddy SR. Profile hidden Markov models. Bioinformatics. 1998;14:755–763. doi: 10.1093/bioinformatics/14.9.755. [DOI] [PubMed] [Google Scholar]
- 20.Baldi P, Chauvin Y, Hunkapiller T, McClure MA. Hidden Markov models of biological primary sequence information. Proc. Natl Acad. Sci. USA. 1994;91:1059–1063. doi: 10.1073/pnas.91.3.1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pugalenthi G, Suganthan PN, Sowdhamini R, Chakrabarti S. Smotif: a server for structural motifs in proteins. Bioinformatics. 2007;23:637–638. doi: 10.1093/bioinformatics/btl679. [DOI] [PubMed] [Google Scholar]
- 22.Sandhya S, Pankaj B, Govind MK, Offmann B, Srinivasan N, Sowdhamini R. CUSP: an algorithm to distinguish structurally conserved and unconserved regions in protein domain alignments and its application in the study of large length variations. BMC Struct. Biol. 2008;8:28. doi: 10.1186/1472-6807-8-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sandhya S, Rani SS, Pankaj B, Govind MK, Offmann B, Srinivasan N, Sowdhamini R. Length variations amongst protein domain superfamilies and consequences on structure and function. PloS One. 2009;4:e4981. doi: 10.1371/journal.pone.0004981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Deepak B, Jun H, Jan P, Jack S, Wei W, Alexander T. Identification of family- specific residue packing motifs and their use for structure-based protein function prediction: Method development. J. Comput. Aided Mol. Des. 2009;23:773–784. doi: 10.1007/s10822-009-9273-4. [DOI] [PubMed] [Google Scholar]
- 26.Yong Lee C, Illarionov B, Woo Y-E, Kemter K, Kim R-R, Eberhardt S, Cushman M, Eisenreich W, Fischer M, Bacher A. Ligand binding properties of the N-terminal domain of Riboflavin Synthase from Escherichia coli. J. Biochem. Mol. Biol. 2007;40:239–246. doi: 10.5483/bmbrep.2007.40.2.239. [DOI] [PubMed] [Google Scholar]
- 27.Winfried M, Sabine E, Adelbert B, Rudolf L. The Structure of the N-terminal Domain of Riboflavin Synthase in Complex with Riboflavin at 2.6 A° Resolution. J. Mol. Biol. 2003;331:1053–1063. doi: 10.1016/s0022-2836(03)00844-1. [DOI] [PubMed] [Google Scholar]
- 28.Mayr G, Domingues FS, Lackner P. Comparative analysis of protein structure alignments. BMC Struct. Biol. 2007;7:50. doi: 10.1186/1472-6807-7-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fischer M, Bacher A. Biosynthesis of vitamin B2: structure and mechanism of riboflavin synthase. Arch. Biochem. Biophys. 2008;474:252–265. doi: 10.1016/j.abb.2008.02.008. [DOI] [PubMed] [Google Scholar]