Abstract
PomBase (www.pombase.org) is a new model organism database established to provide access to comprehensive, accurate, and up-to-date molecular data and biological information for the fission yeast Schizosaccharomyces pombe to effectively support both exploratory and hypothesis-driven research. PomBase encompasses annotation of genomic sequence and features, comprehensive manual literature curation and genome-wide data sets, and supports sophisticated user-defined queries. The implementation of PomBase integrates a Chado relational database that houses manually curated data with Ensembl software that supports sequence-based annotation and web access. PomBase will provide user-friendly tools to promote curation by experts within the fission yeast community. This will make a key contribution to shaping its content and ensuring its comprehensiveness and long-term relevance.
INTRODUCTION
The fission yeast Schizosaccharomyces pombe is a well-studied eukaryotic model organism that has been used since the 1950s to obtain valuable insights into diverse eukaryotic biological processes including the cell growth and division cycle, genome organization and maintenance, cell morphology and cytokinesis, signaling and stress responses, chromatin and gene regulation and meiotic differentiation (1). Moreover, since the completion of its genome sequence in 2002 (2), fission yeast has emerged as a prime model for the characterization of processes relevant to human disease and cell biology. A large and active community engages in biological and biomedical research using this model system, routinely applying molecular genetic, cell biological and biochemical techniques. Data from small- and large-scale projects are accumulating rapidly and will increase substantially over the next few years; the literature corpus currently exceeds 9000 publications and grows by about 500 publications per year.
PomBase (http://www.pombase.org) has recently been established to provide user-friendly and standardized access to genomic features and annotations, enabling scientists to assimilate novel findings into their research programs, improve experimental design, support the interpretation of genetic screens, and facilitate the interpretation of functional genomics and systems biology experiments. An accurate and comprehensive set of manual annotations of gene products based on published data lies at the centre of this database, and is supplemented by automatic annotation and information about non-genic features. The PomBase project aims to provide three key resources to the fission yeast community:
Comprehensive and deep curation of the scientific literature;
A software infrastructure to support curation activities; and
A computational infrastructure to integrate data curated from the literature with the genomic sequence, high-throughput data sets, and data from other fungal genomes.
DATA TYPES AND SOURCES
Historically, functional information about the genome and biology of S. pombe has been maintained in a repository hosted by the GeneDB project at the Wellcome Trust Sanger Institute (WTSI) (http://old.genedb.org/genedb/pombe/). This resource is now superseded by PomBase, which has not only inherited the data from GeneDB, but also includes additional curated data types and high-throughput data sets. The major data types available in PomBase are listed in Table 1.
Table 1.
Summary of data types available in PomBase
| Data type | Distinct descriptors | Total annotations | |
|---|---|---|---|
| Sequence Ontology | DNA features | 31 | 25 650 |
| Protein features | 22 | 919 | |
| Gene Ontology | Biological process | 1861 | 14 501 |
| Molecular function | 1405 | 9149 | |
| Cellular component | 543 | 16 101 | |
| Fission yeast phenotype ontology | 342 | 7043 | |
| Gene product descriptions | 4331 | 7048 | |
| Disease associations | 134 | 378 | |
| PSI-MOD(3) protein modification | 22 | 1861 | |
| EC numbers | 520 | 837 | |
| Name descriptions | 186 | 513 | |
| Annotation extensions | 254 | 340 | |
| Annotation status | 7 | 5142 | |
| BioGRID | Genetic interactions | N/A | 13 147 |
| Physical interactions | N/A | 5131 | |
| Curated fission–budding yeast orthologs | N/A | 5210 |
Sequence feature annotation
DNA and protein features are annotated using the Sequence Ontology (SO) (4). Currently, 31 DNA feature terms are used in 25 650 annotations (examples include gene, exon, tRNA, centromere) and 22 protein feature terms for 919 annotations (examples include nuclear localization signal, ER retention signal, DDB box).
Gene Ontology annotation
Gene Ontology (GO) (5,6) terms are assigned to gene products to represent their molecular functions, cellular components (including complexes), and biological processes. PomBase GO annotation data comprises over 33 500 manual annotations and approximately 3500 automatically assigned annotations, using 3800 unique GO terms.
Phenotype annotation
To support the comprehensive and detailed representation of phenotypes, we are developing the Fission Yeast Phenotype Ontology (FYPO), a formal ontology of phenotypes observed in fission yeast. FYPO is a modular ontology that uses several existing ontologies from the Open Biological and Biomedical Ontologies (OBO) collection (7) as building blocks, including the phenotypic quality ontology PATO (8), GO, and Chemical Entities of Biological Interest (ChEBI) (9). Over 7000 existing annotations have been converted from the legacy GeneDB controlled vocabulary to FYPO terms; these annotations will support sophisticated querying, computational analysis, and comparison between different experiments and even between different species.
Genetic and physical interactions
Annotation of genetic and physical interactions are supported using the BioGRID (http://thebiogrid.org) (10,11) annotation format. Existing annotations curated by BioGRID are imported into PomBase, and newly created annotations will be exchanged with BioGRID.
Genome-scale data
PomBase incorporates a wide variety of data sets that can be mapped to the genome, obtained either from internal sources or via externally loaded URLs or data files. Examples of supported data sets include whole genome re-sequencing data, RNA-seq and ChIP-seq data, various microarray data and other high-throughput data types.
USER ACCESS TO POMBASE
A web portal has been developed for access to the PomBase data, which provides pages describing the current state of annotation of the S. pombe genome, items of interest to the community, and, most importantly, a ‘Gene Overview Page’ that summarizes key information about each gene. Embedded in this portal is a genome browser, providing access to genomic context, sequence-based analyses and high-throughput data.
Gene Overview Pages
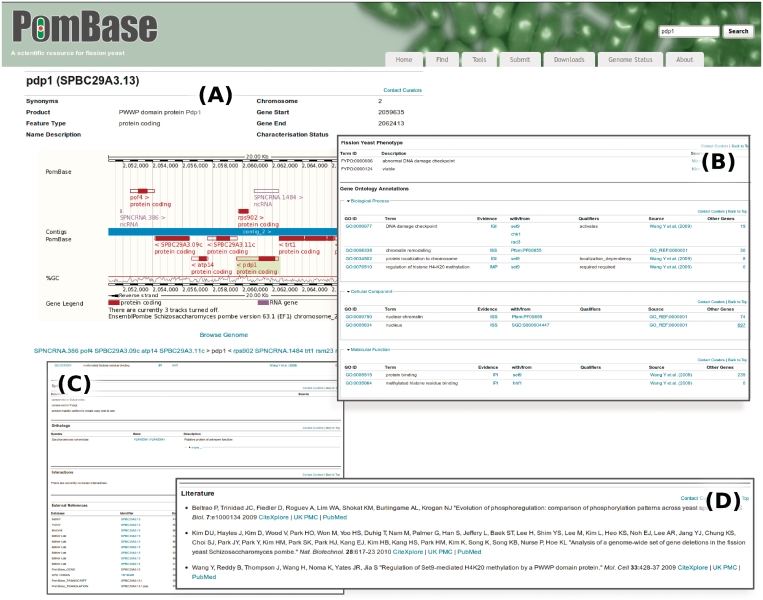
Gene Overview pages organize gene-specific information, including the gene type, product description, sequence features, phenotypes, Gene Ontology annotation and protein modifications as well as physical and genetic interactions. These pages are central to PomBase (see Figure 1).
Figure 1.
PomBase Gene Overview Page highlights. (A) Top of page showing essential information and genomic region. (B) Phenotype and GO annotation. (C) Additional curated data, including orthologs, interactions and external references. (D) Literature pertinent to the gene.
Searching
A simple search is available on every PomBase page. The Advanced Search allows users to perform queries on multiple feature types including GO annotation, protein domain, characterization status, species distribution, protein length, etc. A query history summarizes queries and allows them to be edited or combined using union or intersection.
Genome browser
The PomBase genome browser has been implemented using software developed by the Ensembl project (12). The Ensembl genome browser is a powerful system, offering support for the visualization of sequence, functional annotations, alignments, comparative data and polymorphisms. Many of these features are already exploited by PomBase, and others will be used as and when required for the incorporation of new data types. The use of standard technology readily supports comparative analyses with the genomes of other species that are accessible via Ensembl (also see ‘Implementation’ section, below). Comparative analyses with other fungal genomes and with a range of taxonomically diverse genomes are provided using data generated by the Ensembl Genomes project (13). The Ensembl API enables users interested in comparative analyses to retrieve all data of interest from multiple species present in Ensembl.
IMPLEMENTATION
The implementation of PomBase harnesses and integrates three complementary, well supported and mature technologies, Chado (14), Ensembl and Drupal (http://www.drupal.org). Chado provides an environment for manual curation and the management of curated data while Ensembl provides end-user access via the web portal and display of sequence-based features.
Chado
Curated PomBase data are stored in a PostgreSQL database using the Generic Model Organism Database (GMOD)-compliant Chado schema. Chado supports the management and storage of sequence annotation and literature curation using any combination of available ontologies, and is therefore easily extensible to new data types. Sequence features are curated in Chado using Artemis (15,16). New annotations produced by the PomBase curation team and the fission yeast community, and external data from BioGRID and UniProt/GOA (17), are loaded at regular intervals.
Ensembl
Ensembl is a generic software platform for the automatic annotation, analysis and display of genomes, in use for over a decade. In PomBase, Ensembl provides public access to the integrated fission yeast data, and has been extended to display the deep literature curation managed in Chado. Gene model and annotation data are retrieved from the Chado database using the Bio::Chado::Schema Perl API, and loaded into an Ensembl MySQL database using the Ensembl Perl API. Data in the Ensembl MySQL database are accessible directly or via the Ensembl Perl API, and provide the content served both in the Genome Browser and in the Gene Overview pages. Data from the Ensembl database are also loaded into a BioMart data warehouse (18,19) which supports data mining via web and programmatic interfaces and also provides support for the advanced search (see below).
Drupal
Drupal is a content management system that has been used to provide the web-based portal for PomBase. The use of Drupal has allowed the creation of a clean and intuitive user interface to access information about S. pombe and the PomBase project, and to support community-based functionality including wikis and discussion forums. To support the PomBase interface, two custom Drupal modules have been developed. The Gene Overview module is responsible for generating the Gene Overview pages by retrieving data about specific genes from a custom web service running on the Ensembl web server, which in turn uses the Ensembl Perl API to query the Ensembl MySQL databases. The Query Builder module supports the advanced search interface, and generates and submits custom queries to the BioMart web service to find genes matching specified criteria.
INTEGRATION OF USER DATA
PomBase welcomes contributions from the community to improve the coverage and accuracy of its data. Users wanting to add or modify data in PomBase can directly contact the curation staff (E-mail: helpdesk@pombase.org).
Following a successful pilot project conducted in 2008, a generic web-based curation environment is being developed to support the launch of a comprehensive community curation initiative early in 2012 (Rutherford et al., in preparation). This will allow expert users to directly contribute annotations based on their publications, and will enhance the efforts of core curation staff and contribute to the sustainability of the curation effort in the face of increasing volumes of highly specialized published data.
Additionally, there are many ways that users can directly visualize sequence-based data within the context of the web browser, including access to a Distributed Annotation System, data upload to a private area of the site, and dynamic integration of locally stored BAM files (using standard protocols such as HTTP, allowing users to directly visualize large-scale experimental results in the context of the reference annotation). Producers of mature data sets of any scale that are ready for full integration in PomBase and public dissemination should contact PomBase at the address above.
AVAILABILITY AND DATA PROPAGATION
All of the tools, protocols and workflows developed by PomBase are publicly available (http://www.pombase.org/downloads) and can be implemented by other research communities to create analogous organism-specific databases either in collaboration with the Ensembl Genomes project or independently. Sequence, features and other annotation are available for bulk download via FTP, while subsets of data can be selectively downloaded using the PomBase BioMart.
FUTURE DIRECTIONS
PomBase will continue to incorporate large-scale data sets and curate new data types. We will also incorporate sequence data, automatic annotation, and high-throughput data sets available for other species in the Schizosaccharomyces genus (20).
We anticipate that usage of PomBase will extend beyond the S. pombe community to encompass evolutionary biologists studying genome variations and the evolution of yeasts, fungi, and the eukaryota in general; researchers seeking well-studied orthologs of genes of interest in human and other species; curators from other databases; and bioinformaticians and theoretical biologists requiring programmatic access to fission yeast data in order to construct and test novel hypotheses.
FUNDING
Wellcome Trust [WT090548MA to SGO]. Funding for open access charge: Wellcome Trust.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors thank members of the Ensembl and Ensembl Genomes teams for contributions to the PomBase data pipelines. We also thank Chris Mungall for helpful discussions on Chado and GO, and we thank members of the S. pombe research community whose feedback has helped establish priorities for PomBase development.
REFERENCES
- 1.Egel R, editor. The Molecular Biology of Schizosaccharomyces pombe. Berlin, Germany: Springer; 2004. [Google Scholar]
- 2.Wood V, Gwilliam R, Rajandream M-A, Lyne M, Lyne R, Stewart A, Sgouros J, Peat N, Hayles J, Baker S, et al. The genome sequence of Schizosaccharomyces pombe. Nature. 2002;415:871–880. doi: 10.1038/nature724. [DOI] [PubMed] [Google Scholar]
- 3.Montecchi-Palazzi L, Beavis R, Binz P-A, Chalkley RJ, Cottrell J, Creasy D, Shofstahl J, Seymour SL, Garavelli JS. The PSI-MOD community standard for representation of protein modification data. Nat. Biotechnol. 2008;26:864–866. doi: 10.1038/nbt0808-864. [DOI] [PubMed] [Google Scholar]
- 4.Eilbeck K, Lewis SE, Mungall CJ, Yandell M, Stein L, Durbin R, Ashburner M. The Sequence Ontology: a tool for the unification of genome annotations. Genome Biol. 2005;6:R44. doi: 10.1186/gb-2005-6-5-r44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. The Gene Ontology Consortium (2000) Gene Ontology: tool for the unification of biology. Nat. Genet., 25, 25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. The Gene Ontology Consortium (2010) The Gene Ontology in 2010: extensions and refinements. Nucleic Acids Res., 38, D331–D335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Smith B, Ashburner M, Rosse C, Bard J, Bug W, Ceusters W, Goldberg LJ, Eilbeck K, Ireland A, Mungall CJ, et al. The OBO Foundry: coordinated evolution of ontologies to support biomedical data integration. Nat. Biotechnol. 2007;25:1251–1255. doi: 10.1038/nbt1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gkoutos GV, Green ECJ, Mallon A-M, Hancock JM, Davidson D. Using ontologies to describe mouse phenotypes. Genome Biol. 2005;6:R8. doi: 10.1186/gb-2004-6-1-r8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.deMatos P, Alcántara R, Dekker A, Ennis M, Hastings J, Haug K, Spiteri I, Turner S, Steinbeck C. Chemical Entities of Biological Interest: an update. Nucleic Acids Res. 2010;38:D249–D254. doi: 10.1093/nar/gkp886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stark C, Breitkreutz B-J, Reguly T, Boucher L, Breitkreutz A, Tyers M. BioGRID: a general repository for interaction datasets. Nucleic Acids Res. 2006;34:D535–D539. doi: 10.1093/nar/gkj109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stark C, Breitkreutz B-J, Chatr-Aryamontri A, Boucher L, Oughtred R, Livstone MS, Nixon J, Auken KV, Wang X, Shi X, et al. The BioGRID Interaction Database: 2011 update. Nucleic Acids Res. 2011;39:D698–D704. doi: 10.1093/nar/gkq1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Flicek P, Amode MR, Barrell D, Beal K, Brent S, Chen Y, Clapham P, Coates G, Fairley S, Fitzgerald S, et al. Ensembl 2011. Nucleic Acids Res. 2011;39:D800–D806. doi: 10.1093/nar/gkq1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kersey PJ, Lawson D, Birney E, Derwent PS, Haimel M, Herrero J, Keenan S, Kerhornou A, Koscielny G, Kähäri A, et al. Ensembl Genomes: extending Ensembl across the taxonomic space. Nucleic Acids Res. 2010;38:D563–D569. doi: 10.1093/nar/gkp871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mungall CJ, Emmert DB, FlyBase Consortium A Chado case study: an ontology-based modular schema for representing genome-associated biological information. Bioinformatics. 2007;23:i337–i346. doi: 10.1093/bioinformatics/btm189. [DOI] [PubMed] [Google Scholar]
- 15.Rutherford K, Parkhill J, Crook J, Horsnell T, Rice P, Rajandream MA, Barrell B. Artemis: sequence visualization and annotation. Bioinformatics. 2000;16:944–945. doi: 10.1093/bioinformatics/16.10.944. [DOI] [PubMed] [Google Scholar]
- 16.Carver T, Berriman M, Tivey A, Patel C, Böhme U, Barrell BG, Parkhill J, Rajandream M-A. Artemis and ACT: viewing, annotating and comparing sequences stored in a relational database. Bioinformatics. 2008;24:2672–2676. doi: 10.1093/bioinformatics/btn529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barrell D, Dimmer E, Huntley RP, Binns D, O'Donovan C, Apweiler R. The GOA database in 2009–an integrated Gene Ontology Annotation resource. Nucleic Acids Res. 2009;37:D396–D403. doi: 10.1093/nar/gkn803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smedley D, Haider S, Ballester B, Holland R, London D, Thorisson G, Kasprzyk A. BioMart–biological queries made easy. BMC Genomics. 2009;10:22. doi: 10.1186/1471-2164-10-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kinsella RJ, Kähäri A, Haider S, Zamora J, Proctor G, Spudich G, Almeida-King J, Staines D, Derwent P, Kerhornou A, et al. Ensembl BioMarts: a hub for data retrieval across taxonomic space. Database. 2011;2011 doi: 10.1093/database/bar030. (doi: 10.1093/database/bar030; epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rhind N, Chen Z, Yassour M, Thompson DA, Haas BJ, Habib N, Wapinski I, Roy S, Lin MF, Heiman DI, et al. Comparative functional genomics of the fission yeasts. Science. 2011;332:930–936. doi: 10.1126/science.1203357. [DOI] [PMC free article] [PubMed] [Google Scholar]