Abstract
VectorBase (http://www.vectorbase.org) is a NIAID-supported bioinformatics resource for invertebrate vectors of human pathogens. It hosts data for nine genomes: mosquitoes (three Anopheles gambiae genomes, Aedes aegypti and Culex quinquefasciatus), tick (Ixodes scapularis), body louse (Pediculus humanus), kissing bug (Rhodnius prolixus) and tsetse fly (Glossina morsitans). Hosted data range from genomic features and expression data to population genetics and ontologies. We describe improvements and integration of new data that expand our taxonomic coverage. Releases are bi-monthly and include the delivery of preliminary data for emerging genomes. Frequent updates of the genome browser provide VectorBase users with increasing options for visualizing their own high-throughput data. One major development is a new population biology resource for storing genomic variations, insecticide resistance data and their associated metadata. It takes advantage of improved ontologies and controlled vocabularies. Combined, these new features ensure timely release of multiple types of data in the public domain while helping overcome the bottlenecks of bioinformatics and annotation by engaging with our user community.
INTRODUCTION
VectorBase is a NIAID-funded Bioinformatics Resource Center (BRC) (1), which focuses on arthropod vectors of human pathogens. Our mission is to support the vector research community by providing access to genome assemblies, genome annotations and high-throughput data. VectorBase is involved in capturing community gene annotations, storing microarray expression studies and more recently population biology data. The collection of experimental and sample-related metadata has been aided through our development of ontologies and controlled vocabularies for vector-specific data, such as field-associated samples, pathogen transmission and insecticide resistance. VectorBase currently hosts nine genomes of which the majority are mosquitoes, reflecting their importance in disease agent transmission. The seven corresponding species are: Anopheles gambiae (three genomes, for the PEST, Mali-NIH and Pimperena colonies), Aedes aegypti, Culex quinquefasciatus, Glossina morsitans, Ixodes scapularis, Pediculus humanus and Rhodnius prolixus. We anticipate hosting genome clusters for a broader group of Anopheline mosquitoes, ticks and other important vector genera such as Glossina and Simulium. Full details about current and future genomes to be hosted by VectorBase can be found at http://www.vectorbase.org/organisms. Here, we highlight improvements and new features, and discuss genomes integrated since the last update (2). All information and data are available from our website at http://www.vectorbase.org.
NEW FEATURES
Release cycles and early release of emerging genomes
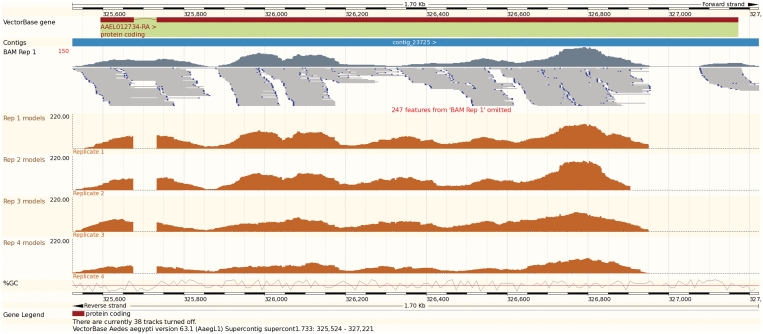
VectorBase now releases data and software updates on a bi-monthly release cycle, such as genome browser improvements via the Ensembl project (3). Recent browser additions include tools for the visualization of user data sources: read coverage plots from high-throughput mRNA-sequencing experiments (BAM (4), WIG http://genome.ucsc.edu/FAQ/FAQformat.html), gene models (GFF3— http://www.sequenceontology.org/gff3.shtml) and population resequencing/variation data sets [VCF (5)] (Figure 1). Searching and selection of evidence tracks have been simplified with a greater level of customization of genome-based views.
Figure 1.
Visualization of user data in the genome browser (image exported directly from the browser). The dark red boxes at the top represent the exons on transcript AAEL012734-RA, and the blue bar represents the contig sequence. The top track in dark grey represents the coverage plot of short-read alignments of an RNA-Seq experiment on the Aedes aegypti transcriptome. Individual read alignments extracted from a BAM-formatted file (4) are displayed underneath in light grey. The four orange tracks represent reconstructed transcript models in four replicates of the same experiment, using a custom RNA-Seq analysis procedure. The BedGraph format was used to generate the data to keep the coverage information base by base.
To make emerging genome sequences rapidly available to our communities, we have recently introduced preliminary sites, called pre-sites, for newly assembled genomes. These contain temporary, unarchived automated gene predictions and transcriptome and proteome alignments. These pre-sites improve vector community involvement during initial analysis, including highly valued community-aided annotation. Once an annotation is finalized, additional analyses are performed such as our standard orthology/paralogy relationship predictions (6) and cross-referencing to other resources. This system was trialled for the R. prolixus and G. morsitans genomes.
Integration of community data
VectorBase has a mandate to capture community annotations. Community appraisal of the reference genome annotations has been important to assess automatic gene predictions and ensure correct models for many gene families as part of the initial genome publication (7) and subsequent analyses (8). Most current annotation data correspond to specific genes and/or gene families and are provided by community members through a simple spreadsheet submitted to our Community Annotation Pipeline. Integration of these data with existing gene sets has greatly improved reference gene sets (e.g. An. gambiae) and has led to a new ‘patch’ build system that uses heuristics to merge manual and automated gene predictions to allow more frequent gene set updates. Patch builds for three species (Ae. aegypti, C. quinquefasciatus and I. scapularis) were performed in 2011. To ensure timely release of community-sourced annotations, all community manual annotation data are made available as a Distributed Annotation System track within the genome browser (9). These data include corrections of gene structures and relevant metadata such as gene symbols and citations. Community-generated transcriptome data from newer sequencing technologies, known as RNA-Seq, are also increasingly being produced for VectorBase species. We have been using these data to validate existing gene models and predict new ones. Alignment algorithms such as Tophat (10), GSNAP (11) (short reads) or GMAP (12) (long reads), were used to map reads to the assembly and identify splicing junctions. Gene models were then reconstructed using Cufflinks (13) and a custom pipeline.
Accessing data
VectorBase has improved its text-based search facility by increasing the speed and the scope of the underlying engine. Search terms now include gene identifiers and descriptions, microarray experiments and expression data. Indices are regenerated for each release using the open source Apache Lucene technology (http://lucene.apache.org) and served using a web service. Information can be retrieved from the search box on the main site or from the genome browser; results contain hyperlinks to genes, their locations and where appropriate, their paralogs/orthologs. A custom interface, CVSearch, has been developed to search (keywords or identifiers) and browse ontologies and controlled vocabularies. More recently, we have used our GDAV open source tool (http://www.vectorbase.org/Help/GDAV) to provide access to available RNA-Seq data. For example, assembled RNA-Seq data for eight Anopheline species for which the genome sequencing is in progress are already available for download or blast, and searchable using keywords, gene identifiers or InterPro domains.
NEW DATA
Ontologies
VectorBase continues to develop and maintain ontologies relating to control of disease vectors (14). Specifically, we host anatomy ontologies [TGMA for mosquitoes and TADS for ticks (15)] and a BFO compliant ontology of insecticide resistance [MIRO (16)]. Our most recent ontology is an extension of the Infectious Disease Ontology (IDO) called IDOMAL (17), which is a comprehensive malaria-focused ontology with more than 2300 unique terms including most related to the disease vector (e.g. vector control). All VectorBase ontologies strictly follow the rules established by the OBO Foundry (18), and can be browsed either at VectorBase or the NCBO Bioportal (http://bioportal.bioontology.org). These ontologies have also been deposited into the publicly accessible OBO Foundry (http://www.obofoundry.org).
Insecticide resistance data
IRbase is a dedicated section of VectorBase that hosts data from both published studies and recently analyzed data for field populations. It used to depend on our MIRO ontology but now relies on the newer IDOMAL ontology described above. We are in the process of incorporating these data into the population biology resource described in the next section.
Variation data
As anticipated in our previous update (2), analyses of populations and variations at the genomic level have increased significantly. To accommodate these data sets, VectorBase has continued to improve its Ensembl-based genome browser for visualizing genomic variation data. As of 2011, the current resource contains data from the dbSNP database (19), variations derived from the An. gambiae Mali-NIH (M molecular form) and Pimperena (S molecular form) sequencing project (20), and genotypes obtained with the AgSNP01 SNP-array (21). We expect to increasingly use this functionality with the completion of a number of planned large-scale population sampling projects.
POPULATION GENOMICS RESOURCE
Integral to handling both genomic variations and insecticide resistance data is the capture of metadata, such as field collection locations and methods. The original IRbase (16) and more recent AgPopGenBase data from UC Davis/UCLA (http://www.vectorbase.org/PopulationData) were highly valuable but were not designed to store more diverse data types. To allow more flexibility, we developed a unified population biology resource that can store all of these data while linking to the genome browser when useful, e.g. high-throughput genotyping data from stored AgSNP01 chip hybridizations (21). This new resource currently contains just over 15 000 mosquito samples originating from over 1600 field collections and more than 34 000 phenotype/genotype assay results.
Population genomics database
We participated in the development of a Chado Natural Diversity Module (22) in collaboration with the GMOD consortium (http://gmod.org) and specific members (23–25). This module is an extension to the Chado database schema that stores population and variation data. The module has a simple, ontology-centred, design which allows the processing of data from a wide range of experiments by extending existing ontologies or adopting new ones.
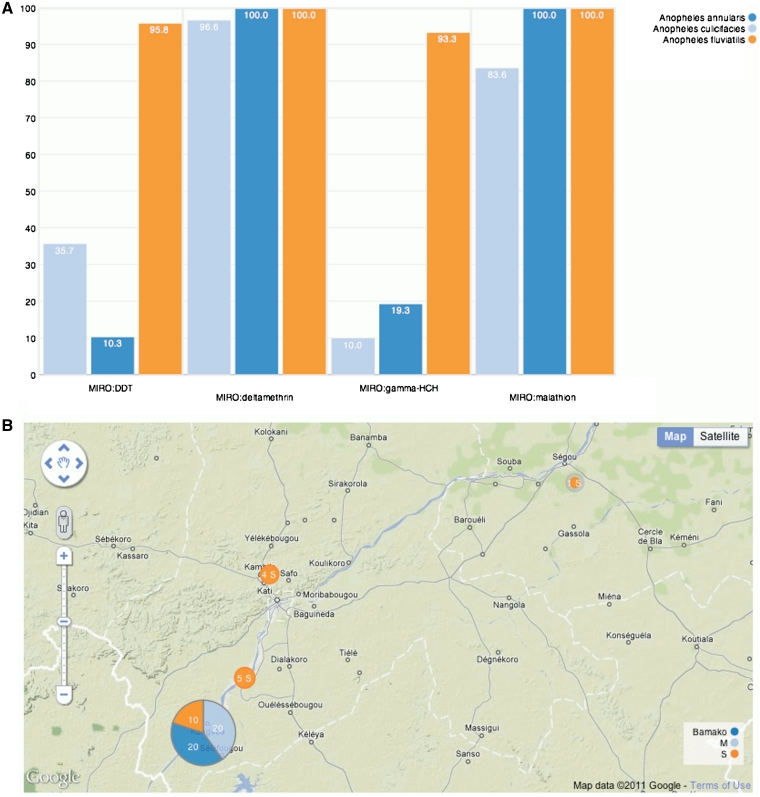
Data storage and access is simplified through Perl and Ruby Application Programming Interfaces (APIs). The Ruby API has been used to write a ‘RESTful’ web service that enables programs, both within VectorBase and from third parties, to retrieve data from the database in a structured format (JSON). The web service code is available under an open source license (http://www.vectorbase.org/Tools). For display of these data, we have developed a lightweight browser and JavaScript library; this queries the main data server and formats it using a set of standard display methods (Figure 2). Display code is available under a GPLv3 license from the same URL as the web service code.
Figure 2.
Examples of customizable displays from the Phenovis javascript library. (A) Susceptibility status of Anopheles fluviatilis, An. annularis and An. culicifacies to insecticides in Koraput District, Orissa: [insecticide x per cent mortality]. (B) Anopheles gambiae M, S and Bamako populations [location x population] (21).
Community-led development
The standard display methods provide a wide variety of options that can be customized by a submitter to best suit their data. By using an open web service and providing the visualization code under an open source license, we hope third-party displays will be developed and we will support these efforts through outreach and through VectorBase-hosted development mailing lists. As a concrete example, we have tested a number of visualizations that retrieve data from our resource and from the web service at EuPathDB (26). Other examples of this approach include the display of climatic, economic or human disease data. This functionality could enable co–analysis of vector and pathogen data of this kind.
Data submission
Data can be submitted to the VectorBase Population Biology Resource via spreadsheet forms using open source tools to assist with formatting and ontology term selection (ISA-Tab (27) and Phenote, http://www.phenote.org). Genotypes are submitted to the variation resource in standard VCF format (5).
EXPANDING THE TAXONOMIC COVERAGE OF VECTORBASE
The decreasing cost of genome sequencing has radical effects on the scope of genome projects. Previously, VectorBase has partnered with large-scale sequencing centres to generate annotation and support single representatives from important vector genera, e.g., An. gambiae for Anopheles and Ae. aegypti for Aedes. Projects using newer generation sequencing methodologies can deliver assemblies at a fraction of the cost and have expanded to encompass multiple species from each genera. NIAID/NHGRI has approved several of these genome clusters including 15 Anopheline genomes, 11 Simulium genomes, 5 Glossina genomes, 2 tick genomes (including the improvement of the I. scapularis assembly) and a mite genome. In total, these represent a 4-fold increase of the number of genomes stored in VectorBase.
VectorBase will support these expanded genome clusters using many of the features described in this update. Each project will produce other data types such as RNA-Seq and variation data through population sampling. VectorBase has also developed a new genome annotation pipeline to infer gene structures from closely related orthologs via whole-genome alignment techniques. Thus a single, high-quality reference annotation set can be used to rapidly predict genes in the other members of a genome cluster. The improvements in the storage and visualization of RNA-Seq and variation data will be invaluable for supporting and augmenting these new genomes for our users.
FUTURE DEVELOPMENTS
In this update, we described improvements to existing features and integration of new data. Two significant advancements are the development of a bi-monthly release and pre-sites, providing the latest data at an early stage of their analysis, thus ensuring high community involvement. VectorBase also assists the community with a helpdesk system, on-line help (FAQs, forum, tutorials) and outreach at conferences. Decreasing sequencing costs are producing a wealth of vector-focused genomics data and expanding the taxonomic coverage far beyond mosquitoes. Although a first cluster of 15 Anopheline genomes is being sequenced, three clusters of related non-mosquito vectors are next in line. Re–sequencing or sequencing of individuals from the same species for population genetics study is also becoming more common. The future of vector genomics appears to be an expansion of both taxonomic coverage (breadth) and within-species re-sequencing (depth). By continuously improving its resources, as has been done in the past years, VectorBase is in a good position to meet this exciting challenge.
FUNDING
National Institutes of Health/National Institute for Allergy and Infectious Diseases (grant numbers HHSN266200400039C, HHSN272200900039C); partial support from: the Evimalar network of excellence (grant number 242095); INFRAVEC from the FP7 program of the European Commission (grant number 228421); Transmalariabloc from the FP7 program of the European Commission (grant number HEALTH-F3-2008-223736). Funding for open access charge: National Institutes of Health/National Institute for Allergy and Infectious Diseases [grant number HHSN272200900039C].
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We would like to acknowledge the reviewers for their useful comments and the many researchers that have provided data to our community resources (gene annotations, expression, variation data) and provided feedback.
As well as the authors listed above, the VectorBase Consortium is composed of: The VectorBase Consortium is composed of: European Bioinformatics Institute, UK: Ewan Birney, Martin Hammond, Paul Kersey, Nick Langridge; Harvard University, USA: Kathy S. Campbell, Madeline Corby, David Emmert, William M. Gelbart, Pinglei Zhou; Imperial College London, UK: George K. Christophides, Fotis C. Kafatos; University of California – Davis, USA: Travis Collier, Gregory C. Lanzaro, Yoosook Lee, Charles E. Taylor; University of New Mexico, USA: Phillip Baker, Margaret Werner-Washburne; University of Notre-Dame, USA: Nora J. Besansky, Ryan Butler, Rory Carmichael, David Cieslak, Nathan Konopinski, Andrew Thrasher, Gregory Madey and Frank H. Collins.
REFERENCES
- 1.Greene JM, Collins F, Lefkowitz EJ, Roos D, Scheuermann RH, Sobral B, Stevens R, White O, Di Francesco V. National Institute of Allergy and Infectious Diseases Bioinformatics Resource Centers: new assets for pathogen informatics. Infect. Immun. 2007;75:3212–3219. doi: 10.1128/IAI.00105-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lawson D, Arensburger P, Atkinson P, Besansky NJ, Bruggner RV, Butler R, Campbell KS, Christophides GK, Christley S, Dialynas E, et al. VectorBase: a data resource for invertebrate vector genomics. Nucleic Acids Res. 2009;37:D583–D587. doi: 10.1093/nar/gkn857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Flicek P, Amode MR, Barrell D, Beal K, Brent S, Chen Y, Clapham P, Coates G, Fairley S, Fitzgerald S, et al. Ensembl 2011. Nucleic Acids Res. 2011;39:D800–D806. doi: 10.1093/nar/gkq1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Danecek P, Auton A, Abecasis G, Albers CA, Banks E, DePristo MA, Handsaker RE, Lunter G, Marth GT, Sherry ST, et al. The variant call format and VCFtools. Bioinformatics. 2011;27:2156–2158. doi: 10.1093/bioinformatics/btr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vilella AJ, Severin J, Ureta-Vidal A, Heng L, Durbin R, Birney E. EnsemblCompara GeneTrees: Complete, duplication-aware phylogenetic trees in vertebrates. Genome Res. 2009;19:327–335. doi: 10.1101/gr.073585.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arensburger P, Megy K, Waterhouse RM, Abrudan J, Amedeo P, Antelo B, Bartholomay L, Bidwell S, Caler E, Camara F, et al. Sequencing of Culex quinquefasciatus establishes a platform for mosquito comparative genomics. Science. 2010;330:86–88. doi: 10.1126/science.1191864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Waterhouse RM, Povelones M, Christophides GK. Sequence-structure-function relations of the mosquito leucine-rich repeat immune proteins. BMC Genomics. 2010;11:531. doi: 10.1186/1471-2164-11-531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jenkinson AM, Albrecht M, Birney E, Blankenburg H, Down T, Finn RD, Hermjakob H, Hubbard TJP, Jimenez RC, Jones P, et al. Integrating biological data–the Distributed Annotation System. BMC Bioinformatics. 2008;9(Suppl 8):S3. doi: 10.1186/1471-2105-9-S8-S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu TD, Nacu S. Fast and SNP-tolerant detection of complex variants and splicing in short reads. Bioinformatics. 2010;26:873–881. doi: 10.1093/bioinformatics/btq057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu TD, Watanabe CK. GMAP: a genomic mapping and alignment program for mRNA and EST sequences. Bioinformatics. 2005;21:1859–1875. doi: 10.1093/bioinformatics/bti310. [DOI] [PubMed] [Google Scholar]
- 13.Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Topalis P, Lawson D, Collins FH, Louis C. How can ontologies help vector biology? Trends Parasitol. 2008;24:249–252. doi: 10.1016/j.pt.2008.03.002. [DOI] [PubMed] [Google Scholar]
- 15.Topalis P, Tzavlaki C, Vestaki K, Dialynas E, Sonenshine DE, Butler R, Bruggner RV, Stinson EO, Collins FH, Louis C. Anatomical ontologies of mosquitoes and ticks, and their web browsers in VectorBase. Insect Mol. Biol. 2008;17:87–89. doi: 10.1111/j.1365-2583.2008.00781.x. [DOI] [PubMed] [Google Scholar]
- 16.Dialynas E, Topalis P, Vontas J, Louis C. MIRO and IRbase: IT tools for the epidemiological monitoring of insecticide resistance in mosquito disease vectors. PLoS Negl Trop Dis. 2009;3:e465. doi: 10.1371/journal.pntd.0000465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Topalis P, Mitraka E, Bujila I, Deligianni E, Dialynas E, Siden-Kiamos I, Troye-Blomberg M, Louis C. IDOMAL: an ontology for malaria. Malar. J. 2010;9:230. doi: 10.1186/1475-2875-9-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smith B, Ashburner M, Rosse C, Bard J, Bug W, Ceusters W, Goldberg LJ, Eilbeck K, Ireland A, Mungall CJ, et al. The OBO Foundry: coordinated evolution of ontologies to support biomedical data integration. Nat. Biotechnol. 2007;25:1251–1255. doi: 10.1038/nbt1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sayers EW, Barrett T, Benson DA, Bolton E, Bryant SH, Canese K, Chetvernin V, Church DM, DiCuccio M, Federhen S, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2011;39:D38–D51. doi: 10.1093/nar/gkq1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lawniczak MKN, Emrich SJ, Holloway AK, Regier AP, Olson M, White B, Redmond S, Fulton L, Appelbaum E, Godfrey J, et al. Widespread divergence between incipient Anopheles gambiae species revealed by whole genome sequences. Science. 2010;330:512–514. doi: 10.1126/science.1195755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Neafsey DE, Lawniczak MKN, Park DJ, Redmond SN, Coulibaly MB, Traoré SF, Sagnon N, Costantini C, Johnson C, Wiegand RC, et al. SNP genotyping defines complex gene-flow boundaries among African malaria vector mosquitoes. Science. 2010;330:514–517. doi: 10.1126/science.1193036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jung S, Menda N. The Chado Natural Diversity module: A new generic schema for large-scale phenotyping and genotyping data. Database. 2011 doi: 10.1093/database/bar051. doi:10.1093/database/bar051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bombarely A, Menda N, Tecle IY, Buels RM, Strickler S, Fischer-York T, Pujar A, Leto J, Gosselin J, Mueller LA. The Sol Genomics Network (solgenomics.net): growing tomatoes using Perl. Nucleic Acids Res. 2011;39:D1149–D1155. doi: 10.1093/nar/gkq866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jaiswal P. Gramene database: a hub for comparative plant genomics. Methods Mol. Biol. 2011;678:247–275. doi: 10.1007/978-1-60761-682-5_18. [DOI] [PubMed] [Google Scholar]
- 25.Jung S, Staton M, Lee T, Blenda A, Svancara R, Abbott A, Main D. GDR (Genome Database for Rosaceae): integrated web-database for Rosaceae genomics and genetics data. Nucleic Acids Res. 2008;36:D1034–D1040. doi: 10.1093/nar/gkm803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aurrecoechea C, Brestelli J, Brunk BP, Fischer S, Gajria B, Gao X, Gingle A, Grant G, Harb OS, Heiges M, et al. EuPathDB: a portal to eukaryotic pathogen databases. Nucleic Acids Res. 2010;38:D415–D419. doi: 10.1093/nar/gkp941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rocca-Serra P, Brandizi M, Maguire E, Sklyar N, Taylor C, Begley K, Field D, Harris S, Hide W, Hofmann O, et al. ISA software suite: supporting standards-compliant experimental annotation and enabling curation at the community level. Bioinformatics. 2010;26:2354–2356. doi: 10.1093/bioinformatics/btq415. [DOI] [PMC free article] [PubMed] [Google Scholar]