Abstract
Hot spots are energetically important residues at protein interfaces and they are not randomly distributed across the interface but rather clustered. These clustered hot spots form hot regions. Hot regions are important for the stability of protein complexes, as well as providing specificity to binding sites. We propose a database called HotRegion, which provides the hot region information of the interfaces by using predicted hot spot residues, and structural properties of these interface residues such as pair potentials of interface residues, accessible surface area (ASA) and relative ASA values of interface residues of both monomer and complex forms of proteins. Also, the 3D visualization of the interface and interactions among hot spot residues are provided. HotRegion is accessible at http://prism.ccbb.ku.edu.tr/hotregion.
INTRODUCTION
Proteins interact with other proteins through their interfaces in order to fulfill their functions. Interfaces are formed by residues whose properties determine binding specificity and affinity. Correct orientations of the residues are critical for complex formation. Interactions between the residues in the binding sites are higher than the protein surface which shows that protein–protein interactions are highly depending on the cooperativity of the residues (1).
Some proteins interact with one or two proteins. Some other proteins, called hub proteins, may interact with many proteins as many as tens of other proteins. It is physically impossible for these hub proteins to interact with all its partners at the same time, since the surface area of the hub protein is fixed. This suggests that there are binding sites that should be used repeatedly to bind different proteins (2–4), probably each with different affinity and specificity. The distribution of the residues across the interface and the residue–residue interactions may answer the question ‘How can the interfaces recognize their partners?’. The residues tend to behave cooperatively during the interactions and they form modules in the interface (5). Proteins may utilize these modules in order to have specificity and affinity during interactions (6–9) and also the combinations of these modules yield a powerful mechanism for binding multiple partners via unique interfaces (10,11). Also, Chakrabarti and Janin (12) stated that small binding sites have single continuous patch; however, larger interfaces may have several patches. Previously, modules in interfaces are defined with various methods such as (i) the edge betweenness criteria in the residue–residue interaction network across the interface (7,13), (ii) difference of energy profiles of residues in interfaces (6,14,15), and (iii) clustering of structurally conserved residues in interfaces (9,11,16). In the edge betweenness approach, the authors used the topology of the network without considering residue energy profiles. The other two approaches used hot spot residues which are driven by energy profiles or structural conservation of residues. The residues that contribute more to the binding free energy are called ‘hot spots’ (17–19). Hot spots are tightly packed and structurally conserved residues (9,11,16). Also Keskin et al. (9) showed that these hot spot residues are not randomly distributed along the protein–protein interfaces; rather clustered. Besides, there is a correlation between energy change and decrease in the accessible surface area of these hot spots (20). Also, the cooperativity of the hot spot residues enlightens the complex binding organizations of the protein–protein interfaces (21,22,23). Computational methods (24–30) are widely used to extract hot spot information from interface, because experimental studies are available for a very limited number of complexes.
In this work, we combine the residue network topology with the residue energy profile based clustering approaches. The residue clusters in interfaces are called ‘hot regions’ (9,22). As we showed in our previous study (22), hot regions are useful to interpret the protein interface properties. Here, we present our database ‘HotRegion’ in order to illustrate hot spot cooperativity information at protein–protein interfaces. HotRegion stores all available protein–protein interfaces which are extracted from Protein Data Bank (PDB) (31) entries using a dynamic update system which is based on the user’s search queries. If a user searches for hot regions via a PDB ID which is not in the HotRegion database, the database can rapidly update itself and show the results. We hope the database will help in detecting cooperativity of functionally important residues, mutagenesis targets and understand the stability and specificity of protein–protein interfaces.
HOTREGION METHOD
An interface is the contact region between two interacting proteins. Two residues are defined to be contacting if the distance between any two atoms of the two residues from different chains is less than the sum of their corresponding van der Waals radii plus 0.5 Å (32,33). Hotspot residues in interfaces are predicted with HotPoint (28) using accessible surface area (ASA) and knowledge based pair energies of each residue (34). In order to define hot regions, a network of hotspots is constructed. In the network, the nodes are the hotspot residues and the edges are linked between nodes if the two hotspot residues are in contact. Two hotspot residues are defined as contacting if the distance between their Cα atoms is smaller than 6.5 Å (9). Afterwards, connected components of the network are found and if the nodes in a connected component are equal or greater than three, the connected component is labeled as a hot region and the hotspot residues in this connected component labeled as the members of this hot region.
DATABASE PROPERTIES
The HotRegion database is available at http://prism.ccbb.ku.edu.tr/hotregion. HotRegion embraces three major components: a relational database management system for data storage and management, a web application to interface the database and a dynamically database update system. Data are stored in a relational MySQL database. The web application runs on an Apache web server hosted on a linux-based system. PHP and JavaScript are used to implement the web application. The database can be updated dynamically.
DATABASE CONTENT
Currently, HotRegion contains all the PDB entries as of January 2011 (70 695 PDB entries, 147 892 protein–protein interfaces). If a user searches hot region information of a protein complex (via PDB ID) which is not in the HotRegion, the database can rapidly update itself and show the results. HotRegion has only protein–protein interface information. HotRegion database offers the researchers to find the hot regions of the protein complexes and provides structural properties of these complexes such as pair potentials of interface residues, ASA and relative ASA values of interface residues of both monomer and complex forms of proteins. Also, the visualization of the interface by using Jmol (35) and residue networks of interactions of hot spot residues are presented in the results. An advanced search option is also available. Users can manipulate the HotRegion parameters by changing default values in advanced search section. Advanced searches are deposited in the database and users can retrieve their jobs by using an email and job id from the ‘Retrieve Job’ section.
HotRegion needs atomic coordinates of the protein complexes in the standard PDB format. If atoms are present in alternative locations, only the first location is considered. For NMR structures, the first model is used. HotRegion is specific to protein–protein interfaces; chains corresponding to DNA and RNA structures return no interface solutions.
If users do not supply enough information, the database asks for the missing information. The HotRegion database is free, open to all users and there are no login requirements.
TUTORIAL
Simple search
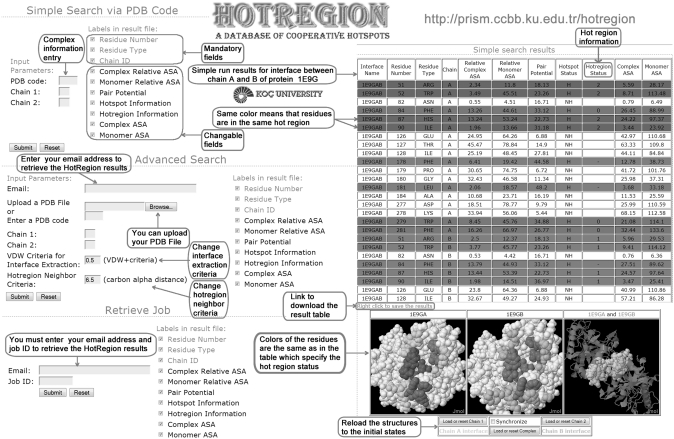
Users retrieve the data of protein interfaces just by entering a PDB ID and two chain identifiers. Between the given monomers there must be an interface in order to get the hot region information. Also users have a control over the presentation of the results. Three properties of the interface (residue number, residue type, chain id) are always displayed in the result table and the output file, and the rest are displayed based on the preferences (Figure 1).
Figure 1.
Properties of HotRegion Database in a quick view.
Advanced search
Users can retrieve the data based on their interface and hot region finding criteria. Users must enter email information in order to retrieve their jobs afterwards. They can supply a PDB file or enter a PDB code. After entering the chain information of the monomers that have an interface between them, users can decide a valid interface extraction threshold which is summed with van der Waals radii of atoms. When the van der Waals threshold gets bigger, the number of interface residues will increase. Also users can change the hot spot neighbor criterion which is the Cα distance between the hot spots. When the hot region criterion gets bigger, the number of hot regions will decrease and hot regions start to merge in order to build larger hot regions.
Retrieve Job
The returning users can retrieve results of previous jobs by using the job ids and their email addresses.
CASE STUDY
Contribution to binding affinity of the proteins
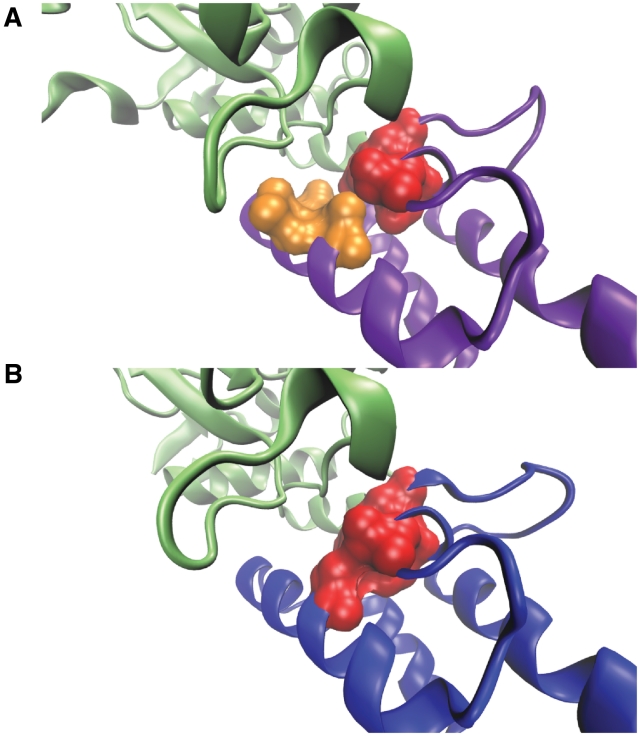
Colicins are plasmid-encoded, stress-induced protein antibiotics that specifically target Escherichia coli cells. When it binds to a specific (cognate) partner, the nuclease can protect the organism from endogenous and incoming colicin (36). Kleanthous and coworkers (37) showed that a limited number of mutations at the interface provide high-affinity binding to a non-cognate partner. According to this work, a non-cognate complex between the colicin E9 endonuclease (E9 DNase) and immunity protein 2 (Im2) (PDB Id: 2WPT) has a weaker binding affinity than the cognate femtomolar E9 DNase—Im9 (PDB Id: 1EMV) interaction. When they substitute three Im2 residues with their Im9 counterparts (Im2 D33L/N34V/R38T) the binding energy is almost similar to the binding energy of cognate complex energy. HotRegion results for these complexes show that the predicted hot spots overlap with the experimental findings. The cognate complex has two hot regions but the non-cognate complex has one hot region (Figure 2) (Table 1). The structural differences at the interface are based on the different side chain orientations. Possibly, cognate complex utilizes the two hot regions at the interface in order to increase the binding affinity of interaction. When we compare the hot regions of both complexes, we observed that the only difference between the hot region residues at the cognate complex are L33 and V34 (they formed the extra hot region with T37 in cognate complex). When these residues are mutated in the non-cognate complex to L and V, these residues may probably form the extra hot region with T37 at non-cognate complex in order to increase the binding affinity of the non-cognate complex as much as the one of the cognate complex.
Figure 2.
(A) Colicin E9 endonuclease (green) interacts with Im9 (purple) and the complex has two hot regions (red and orange). (B) Colicin E9 endonuclease (green) interacts with Im2 (blue) and the complex has one hot region (red).
Table 1.
Hot region results from HotRegion Database for interfaces 1EMVAB and 2WPTAB
| Interface | Residue number | Residue type | Chain | Hot region identifier |
|---|---|---|---|---|
| 1EMVAB | 33 | LEU | A | 1 |
| 1EMVAB | 34 | VAL | A | 1 |
| 1EMVAB | 37 | VAL | A | 1 |
| 1EMVAB | 50 | SER | A | 0 |
| 1EMVAB | 53 | ILE | A | 0 |
| 1EMVAB | 54 | TYR | A | 0 |
| 2WPTAB | 37 | VAL | A | 0 |
| 2WPTAB | 50 | SER | A | 0 |
| 2WPTAB | 53 | ILE | A | 0 |
| 2WPTAB | 54 | TYR | A | 0 |
CONCLUSION
A protein–protein interface consists of two binding sites of two proteins interacting with each other. For all different protein interactions, the binding energies of each complex are miscellaneous and the hot spot residues are distributed in a distinctive pattern. Extracting hot region information from not uniformly distributed binding energy of interfaces is important for analyzing the binding sites of the proteins. Some complexes are built upon more than one hot region, and size of the hot region is changing according to the binding site properties.
We have earlier shown that such hot regions (hotspot clusters) are a signature for the protein–protein interfaces especially for hub proteins (22). A hub protein binds different partner proteins by using different hot regions. These networked hotspot organization may imply that the contribution of the hotspots to the stability of the protein–protein complex within a hot region is cooperative. We hope the database will help in detecting cooperativity of functionally important residues, mutagenesis targets and understand the stability and specificity of protein–protein interfaces.
FUNDING
This project has been supported by TUBITAK (Research Grant No 109T343 and 109E207) and The Turkish Academy of Sciences (TUBA). Funding for open access charge: The open access publication charge for this paper has been waived by Oxford University Press - NAR Editorial Board members are entitled to one free paper per year in recognition of their work on behalf of the journal.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We acknowledge TUBITAK (Research Grant No 109T343 and 109E207) and The Turkish Academy of Sciences (TUBA).
REFERENCES
- 1.Illingworth CJ, Scott PD, Parkes KE, Snell CR, Campbell MP, Reynolds CA. Connectivity and binding-site recognition: applications relevant to drug design. J. Comput. Chem. 2010;31:2677–2688. doi: 10.1002/jcc.21561. [DOI] [PubMed] [Google Scholar]
- 2.Kim PM, Lu LJ, Xia Y, Gerstein MB. Relating three-dimensional structures to protein networks provides evolutionary insights. Science. 2006;314:1938–1941. doi: 10.1126/science.1136174. [DOI] [PubMed] [Google Scholar]
- 3.Keskin O, Nussinov R. Similar binding sites and different partners: implications to shared proteins in cellular pathways. Structure. 2007;15:341–354. doi: 10.1016/j.str.2007.01.007. [DOI] [PubMed] [Google Scholar]
- 4.Tuncbag N, Gursoy A, Guney E, Nussinov R, Keskin O. Architectures and functional coverage of protein-protein interfaces. J. Mol. Biol. 2008;381:785–802. doi: 10.1016/j.jmb.2008.04.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu T, Whitten ST, Hilser VJ. Functional residues serve a dominant role in mediating the cooperativity of the protein ensemble. Proc. Natl Acad. Sci. USA. 2007;104:4347–4352. doi: 10.1073/pnas.0607132104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reichmann D, Rahat O, Albeck S, Meged R, Dym O, Schreiber G. The modular architecture of protein-protein binding interfaces. Proc. Natl Acad. Sci. USA. 2005;102:57–62. doi: 10.1073/pnas.0407280102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Del Sol A, Arauzo-Bravo MJ, Amoros D, Nussinov R. Modular architecture of protein structures and allosteric communications: potential implications for signaling proteins and regulatory linkages. Genome Biol. 2007;8:R92. doi: 10.1186/gb-2007-8-5-r92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tyagi M, Shoemaker BA, Bryant SH, Panchenko AR. Exploring functional roles of multibinding protein interfaces. Protein Sci. 2009;18:1674–1683. doi: 10.1002/pro.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Keskin O, Ma B, Nussinov R. Hot regions in protein–protein interactions: the organization and contribution of structurally conserved hot spot residues. J. Mol. Biol. 2005;345:1281–1294. doi: 10.1016/j.jmb.2004.10.077. [DOI] [PubMed] [Google Scholar]
- 10.Martin J. Beauty is in the eye of the beholder: proteins can recognize binding sites of homologous proteins in more than one way. PLoS Comput. Biol. 2010;6:e1000821. doi: 10.1371/journal.pcbi.1000821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Keskin O, Ma B, Rogale K, Gunasekaran K, Nussinov R. Protein-protein interactions: organization, cooperativity and mapping in a bottom-up Systems Biology approach. Phys. Biol. 2005;2:S24–S35. doi: 10.1088/1478-3975/2/2/S03. [DOI] [PubMed] [Google Scholar]
- 12.Chakrabarti P, Janin J. Dissecting protein-protein recognition sites. Proteins. 2002;47:334–343. doi: 10.1002/prot.10085. [DOI] [PubMed] [Google Scholar]
- 13.Carbonell P, Nussinov R, del Sol A. Energetic determinants of protein binding specificity: insights into protein interaction networks. Proteomics. 2009;9:1744–1753. doi: 10.1002/pmic.200800425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Humphris EL, Kortemme T. Design of multi-specificity in protein interfaces. PLoS Comput. Biol. 2007;3:e164. doi: 10.1371/journal.pcbi.0030164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moza B, Buonpane RA, Zhu P, Herfst CA, Rahman AK, McCormick JK, Kranz DM, Sundberg EJ. Long-range cooperative binding effects in a T cell receptor variable domain. Proc. Natl Acad. Sci. USA. 2006;103:9867–9872. doi: 10.1073/pnas.0600220103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Halperin I, Wolfson H, Nussinov R. Protein-protein interactions; coupling of structurally conserved residues and of hot spots across interfaces. Implications for docking. Structure. 2004;12:1027–1038. doi: 10.1016/j.str.2004.04.009. [DOI] [PubMed] [Google Scholar]
- 17.Bogan AA, Thorn KS. Anatomy of hot spots in protein interfaces. J. Mol. Biol. 1998;280:1–9. doi: 10.1006/jmbi.1998.1843. [DOI] [PubMed] [Google Scholar]
- 18.Clackson T, Wells JA. A hot spot of binding energy in a hormone-receptor interface. Science. 1995;267:383–386. doi: 10.1126/science.7529940. [DOI] [PubMed] [Google Scholar]
- 19.Wells JA. Systematic mutational analyses of protein-protein interfaces. Methods Enzymol. 1991;202:390–411. doi: 10.1016/0076-6879(91)02020-a. [DOI] [PubMed] [Google Scholar]
- 20.Guharoy M, Chakrabarti P. Conservation and relative importance of residues across protein-protein interfaces. Proc. Natl Acad. Sci. USA. 2005;102:15447–15452. doi: 10.1073/pnas.0505425102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shulman-Peleg A, Shatsky M, Nussinov R, Wolfson HJ. Spatial chemical conservation of hot spot interactions in protein-protein complexes. BMC Biol. 2007;5:43. doi: 10.1186/1741-7007-5-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cukuroglu E, Gursoy A, Keskin O. Analysis of hot region organization in hub proteins. Ann. Biomed. Eng. 2010;38:2068–2078. doi: 10.1007/s10439-010-0048-9. [DOI] [PubMed] [Google Scholar]
- 23.Ahmad S, Keskin O, Mizuguchi K, Sarai A, Nussinov R. CCRXP: exploring clusters of conserved residues in protein structures. Nucleic Acids Res. 2010;38:W398–W401. doi: 10.1093/nar/gkq360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Armon A, Graur D, Ben-Tal N. ConSurf: an algorithmic tool for the identification of functional regions in proteins by surface mapping of phylogenetic information. J. Mol. Biol. 2001;307:447–463. doi: 10.1006/jmbi.2000.4474. [DOI] [PubMed] [Google Scholar]
- 25.Guharoy M, Chakrabarti P. Empirical estimation of the energetic contribution of individual interface residues in structures of protein-protein complexes. J. Comput. Aided Mol. Des. 2009;23:645–654. doi: 10.1007/s10822-009-9282-3. [DOI] [PubMed] [Google Scholar]
- 26.Guney E, Tuncbag N, Keskin O, Gursoy A. HotSprint: database of computational hot spots in protein interfaces. Nucleic Acids Res. 2008;36:D662–D666. doi: 10.1093/nar/gkm813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kortemme T, Baker D. A simple physical model for binding energy hot spots in protein-protein complexes. Proc. Natl Acad. Sci. USA. 2002;99:14116–14121. doi: 10.1073/pnas.202485799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tuncbag N, Keskin O, Gursoy A. HotPoint: hot spot prediction server for protein interfaces. Nucleic Acids Res. 2010;38:W402–W406. doi: 10.1093/nar/gkq323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhu X, Mitchell JC. KFC2: a knowledge-based hot spot prediction method based on interface solvation, atomic density, and plasticity features. Proteins. 2011;79:2671–2683. doi: 10.1002/prot.23094. [DOI] [PubMed] [Google Scholar]
- 30.Shulman-Peleg A, Shatsky M, Nussinov R, Wolfson HJ. MultiBind and MAPPIS: webservers for multiple alignment of protein 3D-binding sites and their interactions. Nucleic Acids Res. 2008;36:W260–W264. doi: 10.1093/nar/gkn185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Keskin O, Tsai CJ, Wolfson H, Nussinov R. A new, structurally nonredundant, diverse data set of protein-protein interfaces and its implications. Protein Sci. 2004;13:1043–1055. doi: 10.1110/ps.03484604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tsai CJ, Lin SL, Wolfson HJ, Nussinov R. A dataset of protein-protein interfaces generated with a sequence-order-independent comparison technique. J. Mol. Biol. 1996;260:604–620. doi: 10.1006/jmbi.1996.0424. [DOI] [PubMed] [Google Scholar]
- 34.Tuncbag N, Gursoy A, Keskin O. Identification of computational hot spots in protein interfaces: combining solvent accessibility and inter-residue potentials improves the accuracy. Bioinformatics. 2009;25:1513–1520. doi: 10.1093/bioinformatics/btp240. [DOI] [PubMed] [Google Scholar]
- 35.Herraez A. Biomolecules in the computer: Jmol to the rescue. Biochem. Mol. Biol. Educ. 2006;34:255–261. doi: 10.1002/bmb.2006.494034042644. [DOI] [PubMed] [Google Scholar]
- 36.Kleanthous C, Walker D. Immunity proteins: enzyme inhibitors that avoid the active site. Trends Biochem. Sci. 2001;26:624–631. doi: 10.1016/s0968-0004(01)01941-7. [DOI] [PubMed] [Google Scholar]
- 37.Meenan NA, Sharma A, Fleishman SJ, Macdonald CJ, Morel B, Boetzel R, Moore GR, Baker D, Kleanthous C. The structural and energetic basis for high selectivity in a high-affinity protein-protein interaction. Proc. Natl Acad. Sci. USA. 2010;107:10080–10085. doi: 10.1073/pnas.0910756107. [DOI] [PMC free article] [PubMed] [Google Scholar]