Abstract
As the relevant literature and the number of experiments increase at a super linear rate, databases that curate and collect experimentally verified microRNA (miRNA) targets have gradually emerged. These databases attempt to provide efficient access to this wealth of experimental data, which is scattered in thousands of manuscripts. Aim of TarBase 6.0 (http://www.microrna.gr/tarbase) is to face this challenge by providing a significant increase of available miRNA targets derived from all contemporary experimental techniques (gene specific and high-throughput), while incorporating a powerful set of tools in a user-friendly interface. TarBase 6.0 hosts detailed information for each miRNA–gene interaction, ranging from miRNA- and gene-related facts to information specific to their interaction, the experimental validation methodologies and their outcomes. All database entries are enriched with function-related data, as well as general information derived from external databases such as UniProt, Ensembl and RefSeq. DIANA microT miRNA target prediction scores and the relevant prediction details are available for each interaction. TarBase 6.0 hosts the largest collection of manually curated experimentally validated miRNA–gene interactions (more than 65 000 targets), presenting a 16.5–175-fold increase over other available manually curated databases.
INTRODUCTION
MicroRNAs (miRNAs) are non-protein coding single-stranded endogenous RNAs (∼22 nt long) that can actively regulate gene expression. miRNAs can silence genes through transcript degradation and/or translation suppression in the case of protein coding genes (1). Almost two decades since the identification of lin-4 in Caenorhabditis elegans, miRNAs are now deemed central to the RNA revolution, exhibiting regulatory control to a multitude of crucial cell functions ranging from apoptosis, cell proliferation and differentiation to cell signalling. miRNAs have now been located in a multitude of organisms including plants, algae, viruses and metazoa (2).
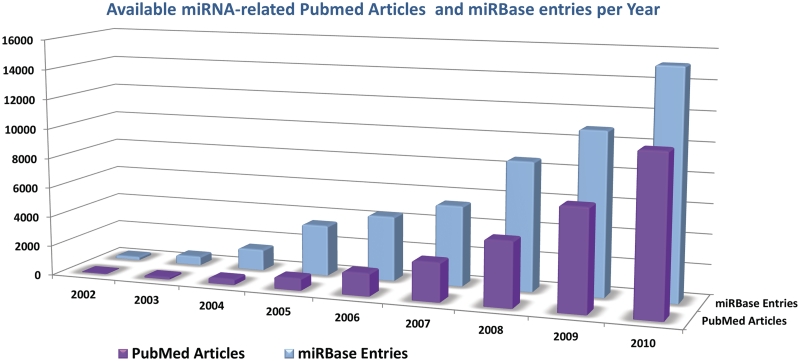
In the past few years, vigorous research efforts have provided significant amounts of data related to miRNA biogenesis and function. This process is reflected in the super linear increase of Sanger Institute's miRBase, the reference database for experimentally supported hairpin precursor and mature miRNA sequences, as well as in the analogous increase of miRNA-related articles in PubMed (Figure 1). The number of experimentally verified miRNAs ranges from a handful to more than a thousand for certain organisms (miRBase, version 17.0); resulting to 1–5% of the number of all protein coding genes in a significant number of cases (2). High-throughput techniques are now playing an important role in miRNA identification (3). Each miRNA is estimated to regulate a large number of targets (4,5). In the case of animals, this number can reach up to several hundreds, resulting in direct miRNA regulation of a remarkably large proportion of the transcriptome (1). The exact mechanisms of this regulation are still debated and are surrounded with substantial controversies. From the initial estimate that miRNA interference was strictly through translation repression in animals and mRNA degradation in plants, we have now arrived at more complicated models that still remain to be further established (1).
Figure 1.
The annual growth of miRNA-related publications in PubMed and the number of entries in miRBase database.
Bioinformatics algorithms and tools are playing a significant role in miRNA target identification. A large number of predictive algorithms are available, which attempt to tackle the problem computationally (2,6). Some targets can be confidently predicted with currently available techniques. However, precision and sensitivity of state-of-the-art algorithms were estimated as ∼50% and 12%, respectively, when tested against proteomics supported miRNA targets (6), highlighting the necessity for mass experimental miRNA target validation (7).
miRNA targets can be experimentally verified with gene-specific, as well as high-throughput techniques. Specific techniques include reporter gene assays, assessment of miRNA and target mRNA co-expression (e.g. northern blotting or qPCR) and estimation of miRNA effect on target protein (e.g. ELISA, western blotting, immunohistochemistry) (8). High-throughput techniques can be a simple extension of an existing gene-specific technique in a high-throughput setting, for example the utilization of microarray screening instead of qPCR. They can also involve novel relevant methodologies, such as RNA-Seq, immunoprecipitation of RISC components, high-throughput sequencing of RNA isolated by crosslinking immunoprecipitation (HITS-CLIP), photoactivatable-ribonucleoside-enhanced crosslinking and immunoprecipitation (PAR-CLIP), biotin tagging of miRNAs, parallel analysis of RNA ends (PARE) and various proteomics approaches such as SILAC (7).
As the relevant literature and the number of experiments increase in a super linear fashion, databases that curate and collect experimentally verified targets have gradually emerged. These databases attempt to provide efficient access to this wealth of experimental data, which is scattered in thousands of manuscripts. TarBase v1.0 (9), the first database for experimental supported miRNA targets was created to assist for the design of more robust prediction algorithms. As the accumulated data increased, TarBase and the other related databases that followed widened their scope of application and became invaluable tools to all facets of miRNA-related research.
AVAILABLE DATABASES FOR EXPERIMENTALLY SUPPORTED TARGETS
A brief overview of the available databases is presented below in alphabetical order.
miR2Disease
miR2Disease (10) was first released in 2008. It is a manually curated database that aims to provide information regarding miRNA-related pathologies. miR2Disease curates 809 miRNA–gene interactions for Homo sapiens, coupled with related disease information derived from relevant literature. The 3273 miRNA disease-related entries consist the strongest point of the database (last updated 14 March 2011). The user can search by miRNA, target gene or disease name. Further details include method of validation, validated targets from an earlier version of TarBase, relation with the pathology, manuscript information and links to target predicting algorithms.
MirnaMAP
MirnaMAP (11) was first released in 2006. It contains data derived from an outdated version of TarBase (346 targets) and by manual curation (29 targets). MirnaMAP has not been updated for >4 years and contains a limited amount of experimentally validated targets for H. sapiens. The largest amount of mirRNAMAP entries is based on predicted interactions for 2464 miRNAs in 12 species. MirnaMAP provides a wealth of available data for each database entry, including miRNA and gene information, bead-array microRNA tissue expression profile, qPCR tissue expression profile, predicted target genes, as well as relevant literature. The 4 years without updates form a substantial drawback.
MiRecords
MiRecords (12) was first released in 2008. It contains manually curated and predicted miRNA targets. The validated targets component of the database contains 2286 interactions between 548 miRNAs and 1579 target genes in nine species (update 25 November 2010). The largest number of those interactions is derived from gene-specific experiments. The database provides miRNA, gene and target site-related information, as well as links to miRBase and RefSeq. miRNA–gene interactions are supported with data regarding manuscript information, experimental method used for validation, as well as a selected passage from the manuscript stating the experimental result. However, the user does not have the ability to filter results based on any of the available predicted or validated component fields. The miRecords interface also enables the user to insert new miRNA–target interactions.
miRSel
miRSel (13) database was first released in 2010. It contains miRNA interaction data derived solely from text mining of MedLine abstracts. The text mining algorithm manages to extract miRNA–gene associations with 65% precision, 90% recall, based on a test performed on 89 selected sentences, derived from 50 PubMed abstracts. MiRSel contains 3690 miRNA–gene text mined associations. By applying less stringent criteria, the user can have access to approximately 8000 pairs, which are deemed as less reliable by the developers. In miRSel, the user can also search for miRNAs related to specific MedLine articles that contain a subset of desired terms or that are related to Gene Ontology entries. Links to external databases such as miRBase and Entrez Gene are provided for each entry. Information regarding the experimental method used for miRNA target validation is not available. Data derived from other curated miRNA interaction databases such as TarBase 5.0, miR2Disease and miRecords have also been integrated.
miRTarBase
miRTarBase (14) was first released in 2010. It includes manually curated data for 3969 experimentally verified miRNA–gene interactions for 14 species (update 15 April 2011). It provides information related to the miRNA, the target gene and the target site. In many cases, where the articles do not explicitly present target site information, miRTarBase can provide predicted regions by using a computational target prediction algorithm. Information regarding available experimental findings supporting the interaction is also included. The user-interface provides links to external data sources such as NCBI Entrez, UCSC Genome Browser, miRBase, BioGPS, iHOP and HGNC. Optionally the user can submit data for non-indexed interactions.
miRWalk
miRWalk (15) was first released in 2010. It provides experimentally supported miRNA targets identified solely from text-mined abstracts available in MedLine. The miRWalk validated targets module hosts text-mined interactions for 1572 miRNAs interacting with 5080 genes for three species (human, mouse and rat). A direct estimation of the system's accuracy in interaction extraction has not been provided. The text mining approach of the authors enabled them to also collect data for disease targets, organs, cell lines and pathways.
StarBase
StarBase (16) was first released in 2010. It is a platform focused on the analysis of high-throughput CLIP-Seq (HITS-CLIP and PAR-CLIP) and degradome sequencing (Degradome-Seq and PARE) data for six organisms. StarBase miRNA-related data are derived from eight different studies. The developers utilized five prediction programs to locate putative targets, which were subsequently intersected with the previously analyzed high-throughput data, resulting in a high number of putative targets (∼500 000). The user can reduce the number of false positives by selecting the results of only one prediction algorithm. StarBase provides the in-house developed deepView Genome Browser, which enables access to mapped reads, predicted and known miRNA targets, ncRNAs, protein coding genes, target clusters, target peaks and target plots. Provided information includes miRNA and gene-related info, GO terms and KEGG pathways related to each target gene.
TarBase 5.0
DIANA Lab's TarBase (17) was first released in 2005 and its fifth version includes 1300 experimentally supported targets from eight species that were manually curated from relevant literature. The provided data for each interaction include miRNA information, target gene general information, the nature of the experimental validation, cell line, sufficiency of the site to induce translational repression and/or cleavage and the supporting manuscript. The database also incorporates functional links to external sources of data for each entry, such as Ensembl, HUGO, UVSV and Swiss-Prot.
As miRNA-related research advances, the requirements for relevant databases increase significantly. An evident necessity exists for databases that manually curate large numbers of experimentally validated miRNA targets from gene-specific as well as from high-throughput techniques. Databases containing both types of experimental data provided until today less than 5000 validated targets, with a significant portion of those targets derived from a few high-throughput experiments. Incorporating a large number of supporting studies increases data validity and enables such databases to become concise datasets for training and testing robust miRNA prediction algorithms. Another important issue which soon became evident was that researchers require advanced searching and result filtering capabilities, in order to accurately discover miRNAs or genes of specific interest. Finally, the enhancement of the datasets with added information from external sources (such as pathways or GO terms) and metadata can enable efficient data mining of the existent experimentally validated results, producing useful novel observations.
The aim of TarBase 6.0 (http://www.microrna.gr/tarbase) is to face the aforementioned current challenges and to inaugurate the next generation of validated miRNA target databases by providing a significant increase of available targets derived from all available experimental techniques, while incorporating a powerful set of tools in a user-friendly interface.
METHODS AND RESULTS
Text mining-assisted literature curation
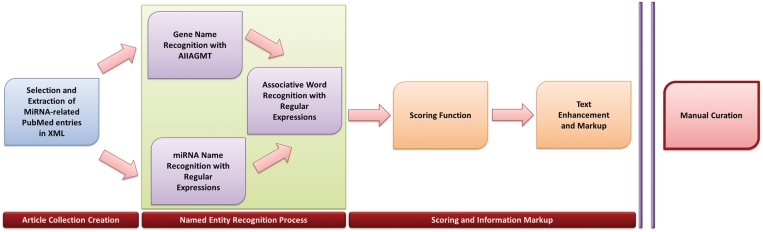
Manual miRNA-related literature curation is a time consuming process that requires highly trained personnel. A text mining-assisted human curation pipeline was implemented, in order to reduce the necessary time for manuscript curation. Since accurate automatic association extraction from biomedical literature is still an open problem, the text mining application was designed at this point only to assist the human curators in their task. A straightforward pipeline was designed that included NER (named entity recognition), miRNA–target association identification, scoring and enhanced text presentation (Figure 2).The designed module provides: markup of genes, miRNAs and associating words with different notations. Subsequently, scoring and sorting of abstracts are performed based on a target existence probability score. Specific markup for sections of increased interest is finally added; for example by underlining sentences that seemed highly probable to include a miRNA–target association.
Figure 2.
The adopted text-mining-assisted curation pipeline.
Initially, all relevant abstracts containing miRNA-related terms (microRNA or micro-RNA or miRNA or ‘micro RNA’) in their title, text, keywords or MesH terms were downloaded from MedLine in the form of XML texts. The subsequent named entity identification process was divided into two distinct steps: in the first step, genes were identified using AIIA Lab's AIIAGMT, one of the highest ranking named entity recognition systems in BioCreative challenges (18). AIIAGMT offers an XML-RPC interface through a Perl module, implemented by the authors, which was incorporated into our pipeline. Our script marked specifically all mentions for genes discovered by AIIAGMT. The recognition of miRNAs is much more straightforward, since they follow a much more conservative nomenclature and do not possess a high number of aliases, which is common ground in gene notation. MiRNA recognition was therefore performed with regular expressions implemented in Perl. Following NER, a list of 16-word stems was used as a basis for associative word recognition within the text. These stems were derived by removing suffixes (from verbs or nouns) from a superset of miRNA–target association denoting words (e.g. target-s, target-ing, target-ed, etc.). Finally, a scoring function was applied that promoted abstracts containing a large number of sentences containing miRNA-target associations. This tool significantly increased daily curation outcome and enabled us to incorporate an unprecedented amount of curated targets in TarBase.
The database
Schema
A new relational database schema was designed and implemented to accommodate present and planned future TarBase updates. TarBase 6.0 hosts a significant amount of information for each miRNA–gene interaction ranging from miRNA and gene-related facts, to information specific to their interaction, the experimental validation methodologies and their subsequent outcomes. All database entries are enriched with a significant amount of function-related data, as well as general information derived from external databases. Database entries are mapped to external sources such as UniProt, Ensembl, RefSeq and others, in order to provide seamless integration with other services. DIANA microT v4 (19) miRNA target prediction scores and links to the relevant microT entry have also been added to the interactions. The new extended database schema was designed to accommodate miRNA–gene interaction data in high detail and to be efficient during query evaluation. The database is supported by a large number of indices and materialized views for performance enhancement.
Data
The database was populated with entries derived from manual manuscript curation. The curators noted the miRNA, the related target gene as well as information regarding the experiment such as the utilized cell line or tissue. The utilized methodology (gene specific or high throughput) was specifically mentioned:
Reporter genes
qPCR
Western blotting
MicroArray
Proteomics (such as pSILAC)
Sequencing (RNA-Seq, HITS-CLIP, PAR-CLIP)
Degradome-Seq
Other (e.g. ELISA, RACE, immunohistochemistry, etc.)
For each entry, the experiment outcome (positive or negative), the type of association (direct or indirect), the type of regulation (up-regulation or down-regulation), the binding site, as well as the outcome for each specific methodology were inserted into the database by the curators. A small excerpt from the manuscript was also added, which was judged to include important information regarding the experiment.
Raw high-throughput data analysis
Raw data sets from high-throughput experiments deposited in relevant repositories or provided in supplements of seven publications were analyzed [four microarray (20–23), two HITS-CLIP (24, 25), one PAR-CLIP (26)]. The analysis methodologies utilized are reported briefly below.
PAR-CLIP and HITS-CLIP data consist of genomic coordinates specifying potential positions of miRNA-binding sites. Each position is scanned for complementary sequence to the seed of all known miRNAs and if such occurrence is found, the gene is noted as targeted by the corresponding miRNA. For the PAR-CLIP data, the seed search is refined by limiting the scanned region within 10 nucleotides around the T to C mutation. The mutation has been shown (26) to occur near the crosslink site of the Ago protein to the mRNA and therefore indicates with higher accuracy the position of the miRNA-binding site. In microarray experiments, all differential expressions exceeding 50%-fold change were included.
Statistics
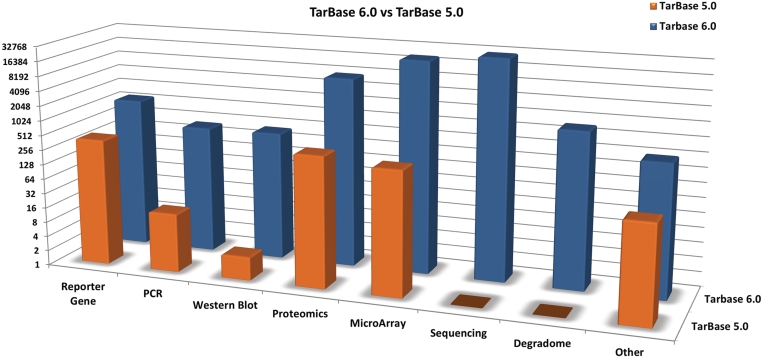
The new database includes 65 814 experimentally validated miRNA–gene interactions, which are extracted from relevant literature by TarBase curators. This is a 50-fold increase of entries from the latest TarBase version and a 16.5- to 175-fold increase from all the other available manually curated databases. The number of entries for each experimental method for TarBase 5.0 and TarBase 6.0 are presented in Figure 3. TarBase 6.0 entries are supported from a large number of manuscripts, significantly increasing data robustness. TarBase 6.0. accommodates a significant number of outcomes procured from state-of-the-art high-throughput studies. Importantly, TarBase hosts data derived from 3 CLIP-Seq and 12 Degradome-Seq studies, which is a 87.5%-fold increase compared to the eight studies supporting StarBase, a database dedicated to collecting data from those methodologies.
Figure 3.
Comparison of the number of targets per experimental validation method of TarBase 5.0 and TarBase 6.0. The number of targets is shown on a log2 scale (i.e. one visible unit represents twice the amount of available targets). Exponential increases thus appear as linear.
Interface
The new TarBase interface attempts to balance ease of use and functionality. Users can browse through numerous miRNA targets and explore the outcomes of hundreds of experimental studies in a simple and intuitive way. However, simplicity should not prevent users from performing complex functions, like processing results using several types of filters or executing combined searches. Such features are usually absent in most previous miRNA target databases. In this section, we present a brief overview of the functionalities provided by the new TarBase interface.
Searching for targets
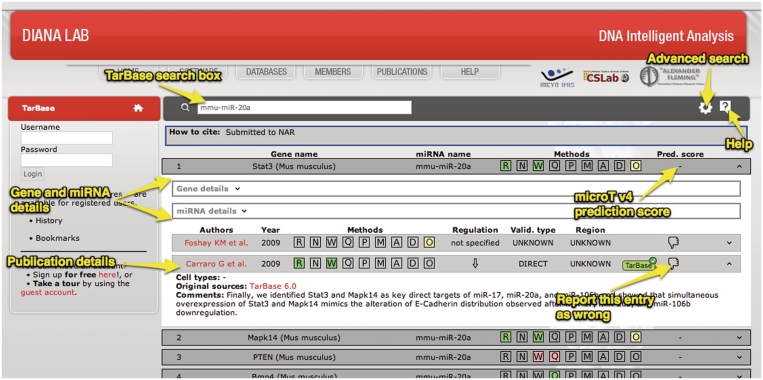
From the TarBase Search Box (Figure 4) users can search for targets by inserting one or more miRNA identifiers, gene identifiers or a combination of miRNAs and genes (combined search). For users’ convenience, there is an extensive support for numerous miRNA and gene identification nomenclatures such as miRNA name or MIMAT id, for miRNAs and gene name, Ensembl id, or RefSeq id for genes. In cases of spelling errors or any other ambiguity in the user's input, the system presents automatic suggestions. In such cases, the user can select any of the options provided to efficiently complete the database query.
Figure 4.
Screenshot of the TarBase 6.0 interface. The results of a query regarding mmu-miR-20a are depicted in the form of an expandable list. The miRNA details section is selected by the user for the first entry, showing details of supporting publications and validation experiments.
Presentation of results
Following the submission of a user query, the system displays related targets and all available relevant information in the form of an enhanced expandable list below the TarBase search box. Each list row corresponds to an experimentally verified miRNA target. Each entry is accompanied with a significant amount of information related to the gene, the miRNA and their interaction. Supporting experimental data are also provided, such as the methods used for validation, the outcome of each method and the DIANA-microT v4 prediction score (19). The prediction score is also hyperlinked to the corresponding microT v4 page, to provide detailed information about the target prediction.
Each entry in the new result interface is expandable, rendering further information in an easy and intuitive way. The user can show or hide this information by simply clicking on the arrows next to each entry. The extra information presented includes (i) detailed description of the involved gene and miRNA (name, description, relevant MeSH terms, transcripts, etc.), (ii) the list of publications that support the interaction, along with details about the experimental methods utilized and their outcomes. Gene and miRNA information is enhanced with relevant KEGG pathways, links to external sources of data such as UniProt and Ensembl, coupled with accession codes to relevant external databases such as RefSeq. On the other hand, interaction data are supported with notations for direct/indirect interaction, up/down-regulation of the target, cell and tissue types used in the experiment and the binding region of the miRNA to the target. Moreover, a PubMed hyperlink to the supporting publication is provided along with an excerpt of the manuscript text denoting useful information regarding each interaction. Finally, tooltips and other means of online help support the TarBase result list, actively assisting user interaction.
Filtering the results
The Advanced Search feature of TarBase interface assists users to customize the result list according to their specific needs. Users can enable this feature by clicking on the gear wheel button at the right of the search box (Figure 4) to activate and display an extensive set of filtering options. Users can also customize the results list by selecting the desired value for each option. Option selection includes (i) species, (ii) experimental validation method (i.e. qPCR, microarray, etc.), (iii) regulation type (i.e. up/down), (iv) type of interaction (i.e. direct/indirect), (v) validation outcome (i.e. positive/negative), (vi) publication year, (vii) DIANA microT prediction score.
Integration of other miRNA target databases
TarBase 6.0 provides the option to the user to access targets indexed in all other manually curated databases. Specifically, TarBase 6.0 integrates entries from miRecords (12), miRTarBase (14) and miR2Disease (10). This functionality is inactive by default but the user can easily enable the integration module from the advanced search menu. Following activation, the records from the external databases are incorporated in the result list. Each entry is accompanied with an explicit mention of database of origin. All statistics mentioned in previous sections regarding the TarBase database did not involve integrated targets or external sources of miRNA interactions but only the in-house curated targets. If the user enables the miRNA target Database Integration module, available TarBase entries increase to 72 794 available entries (11% increase).
Supported and upcoming features
In TarBase 6.0, the user has the ability to denote erroneous entries with the click of a button or submit entries from an experiment by utilizing easy to use active forms. A set of new functionalities is also planned for an upcoming update, including user authentication in order to provide a more personalized user experience, enabling the saving of result sets, queries and the use of customized RSS feeds for alerts of new experimentally validated targets.
CONCLUSION
The transition from TarBase 5.0 to TarBase 6.0 includes a 50-fold target increase, coupled with a significant extension of specific research-oriented features. This update aims at rendering TarBase 6.0 into a multifaceted tool robustly supporting all miRNA-related research. The database will be regularly updated in 4-month intervals in order to capture the growing number of publications covering novel targets. Community support is essential for the success of this process, as has been the case for other curated databases for genes or proteins, where the inclusion of experimental results to a relevant database by the authors is now considered as standard practice. TarBase 6.0 includes an easy to use form for assisted manual entry of miRNA targets and authors are welcome to include their targets following the publication of their experiments. Such practices can increase publication impact as well as the speed and the degree of the dissemination of experimental findings.
FUNDING
Funding for open access charge: Projects 09 SYN - 13 - 1055 ‘MIKRORNA’ and 09 SYN - 13 -901 ‘EDGE’ from the Greek General Secretariat for Research and Technology.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors would like to acknowledge Giorgio Papadopoulos for his substantial contribution in the compilation and updates of TarBase 5.0.
REFERENCES
- 1.Huntzinger E, Izaurralde E. Gene silencing by microRNAs: contributions of translational repression and mRNA decay. Nat. Rev. Genet. 2011;12:99–110. doi: 10.1038/nrg2936. [DOI] [PubMed] [Google Scholar]
- 2.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Friedlander MR, Chen W, Adamidi C, Maaskola J, Einspanier R, Knespel S, Rajewsky N. Discovering microRNAs from deep sequencing data using miRDeep. Nat. Biotechnol. 2008;26:407–415. doi: 10.1038/nbt1394. [DOI] [PubMed] [Google Scholar]
- 4.Baek D, Villen J, Shin C, Camargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Selbach M, Schwanhausser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- 6.Alexiou P, Maragkakis M, Papadopoulos GL, Reczko M, Hatzigeorgiou AG. Lost in translation: an assessment and perspective for computational microRNA target identification. Bioinformatics. 2009;25:3049–3055. doi: 10.1093/bioinformatics/btp565. [DOI] [PubMed] [Google Scholar]
- 7.Thomson DW, Bracken CP, Goodall GJ. Experimental strategies for microRNA target identification. Nucleic Acids Res. 2011;39:6845–6853. doi: 10.1093/nar/gkr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kuhn DE, Martin MM, Feldman DS, Terry AV, Jr, Nuovo GJ, Elton TS. Experimental validation of miRNA targets. Methods. 2008;44:47–54. doi: 10.1016/j.ymeth.2007.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sethupathy P, Corda B, Hatzigeorgiou AG. TarBase: a comprehensive database of experimentally supported animal microRNA targets. RNA. 2006;12:192–197. doi: 10.1261/rna.2239606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jiang Q, Wang Y, Hao Y, Juan L, Teng M, Zhang X, Li M, Wang G, Liu Y. miR2Disease: a manually curated database for microRNA deregulation in human disease. Nucleic Acids Res. 2009;37:D98–D104. doi: 10.1093/nar/gkn714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hsu SD, Chu CH, Tsou AP, Chen SJ, Chen HC, Hsu PW, Wong YH, Chen YH, Chen GH, Huang HD. miRNAMap 2.0: genomic maps of microRNAs in metazoan genomes. Nucleic Acids Res. 2008;36:D165–D169. doi: 10.1093/nar/gkm1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xiao F, Zuo Z, Cai G, Kang S, Gao X, Li T. miRecords: an integrated resource for microRNA-target interactions. Nucleic Acids Res. 2009;37:D105–D110. doi: 10.1093/nar/gkn851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Naeem H, Kuffner R, Csaba G, Zimmer R. miRSel: automated extraction of associations between microRNAs and genes from the biomedical literature. BMC Bioinformatics. 2010;11:135. doi: 10.1186/1471-2105-11-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hsu S-D, Lin F-M, Wu W-Y, Liang C, Huang W-C, Chan W-L, Tsai W-T, Chen G-Z, Lee C-J, Chiu C-M, et al. miRTarBase: a database curates experimentally validated microRNA–target interactions. Nucleic Acids Res. 2011;39:D163–D169. doi: 10.1093/nar/gkq1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dweep H, Sticht C, Pandey P, Gretz N. miRWalk - Database: prediction of possible miRNA binding sites by “walking” the genes of three genomes. J. Biomed. Inform. 2011;44:839–847. doi: 10.1016/j.jbi.2011.05.002. [DOI] [PubMed] [Google Scholar]
- 16.Yang JH, Li JH, Shao P, Zhou H, Chen YQ, Qu LH. starBase: a database for exploring microRNA-mRNA interaction maps from Argonaute CLIP-Seq and Degradome-Seq data. Nucleic Acids Res. 2011;39:D202–D209. doi: 10.1093/nar/gkq1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Papadopoulos GL, Reczko M, Simossis VA, Sethupathy P, Hatzigeorgiou AG. The database of experimentally supported targets: a functional update of TarBase. Nucleic Acids Res. 2009;37:D155–D158. doi: 10.1093/nar/gkn809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smith L, Tanabe LK, Ando RJ, Kuo CJ, Chung IF, Hsu CN, Lin YS, Klinger R, Friedrich CM, Ganchev K, et al. Overview of BioCreative II gene mention recognition. Genome Biol. 2008;9(Suppl. 2):S2. doi: 10.1186/gb-2008-9-s2-s2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maragkakis M, Vergoulis T, Alexiou P, Reczko M, Plomaritou K, Gousis M, Kourtis K, Koziris N, Dalamagas T, Hatzigeorgiou AG. DIANA-microT Web server upgrade supports Fly and Worm miRNA target prediction and bibliographic miRNA to disease association. Nucleic Acids Res. 2011;39:W145–W148. doi: 10.1093/nar/gkr294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gennarino VA, Sardiello M, Avellino R, Meola N, Maselli V, Anand S, Cutillo L, Ballabio A, Banfi S. MicroRNA target prediction by expression analysis of host genes. Genome Res. 2009;19:481–490. doi: 10.1101/gr.084129.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang X. Systematic identification of microRNA functions by combining target prediction and expression profiling. Nucleic Acids Res. 2006;34:1646–1652. doi: 10.1093/nar/gkl068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Linsley PS, Schelter J, Burchard J, Kibukawa M, Martin MM, Bartz SR, Johnson JM, Cummins JM, Raymond CK, Dai H, et al. Transcripts targeted by the microRNA-16 family cooperatively regulate cell cycle progression. Mol. Cell. Biol. 2007;27:2240–2252. doi: 10.1128/MCB.02005-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol. Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chi SW, Zang JB, Mele A, Darnell RB. Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature. 2009;460:479–486. doi: 10.1038/nature08170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zisoulis DG, Lovci MT, Wilbert ML, Hutt KR, Liang TY, Pasquinelli AE, Yeo GW. Comprehensive discovery of endogenous Argonaute binding sites in Caenorhabditis elegans. Nat. Struct. Mol. Biol. 2010;17:173–179. doi: 10.1038/nsmb.1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hafner M, Landthaler M, Burger L, Khorshid M, Hausser J, Berninger P, Rothballer A, Ascano M, Jr, Jungkamp AC, Munschauer M, et al. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell. 2010;141:129–141. doi: 10.1016/j.cell.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]