Abstract
Ensembl Genomes (http://www.ensemblgenomes.org) is an integrative resource for genome-scale data from non-vertebrate species. The project exploits and extends technology (for genome annotation, analysis and dissemination) developed in the context of the (vertebrate-focused) Ensembl project and provides a complementary set of resources for non-vertebrate species through a consistent set of programmatic and interactive interfaces. These provide access to data including reference sequence, gene models, transcriptional data, polymorphisms and comparative analysis. Since its launch in 2009, Ensembl Genomes has undergone rapid expansion, with the goal of providing coverage of all major experimental organisms, and additionally including taxonomic reference points to provide the evolutionary context in which genes can be understood. Against the backdrop of a continuing increase in genome sequencing activities in all parts of the tree of life, we seek to work, wherever possible, with the communities actively generating and using data, and are participants in a growing range of collaborations involved in the annotation and analysis of genomes.
OVERVIEW AND ACCESS
Ensembl Genomes (http://www.ensemblgenomes.org) is organized as five sites, each focused on one of the traditional kingdoms of life: bacteria (specific URL http://bacteria.ensembl.org), protists, fungi, plants and metazoa (invertebrate). Vertebrate metazoa are the focus of the Ensembl project (http://www.ensembl.org) (1); Ensembl Genomes provides a complementary set of interfaces for non-vertebrate species. Core data available for all species includes genome sequence and annotations of protein- and non-coding genes; additional data includes transcriptional data, polymorphisms and comparative analysis. Interactive access is provided through a web interface providing genome browsing capabilities: users can scroll through a graphical representation of a DNA molecule at various levels of resolution, seeing the relative locations of features—including conceptual annotations (e.g. genes, SNP loci), sequence patterns (e.g. repeats) and experimental data (e.g. sequences and external sequence features mapped onto the genome), which often provide direct support for the annotations. Functional information is provided through direct curation, import from UniProt (2), or imputation from protein sequence [using the classification tool InterProScan (3)]. Users can download much of the data available on each page in a variety of formats, and tools exist for upload of (various types of) user data, allowing users to see their own annotation in the context of the reference sequence. A BLAST interface is provided for DNA and protein-based sequence search.
The data are stored in a set of MySQL databases using the same schemas as those in use for the Ensembl project. Direct access to these is provided through a public MySQL server (mysql.ebi.ac.uk:4157; user ‘anonymous’) and additionally through a well-developed Perl API that provides an object-oriented framework for working with the data. Database dumps and common data sets (e.g. DNA, RNA and protein sequence sets, and sequence alignments) can be directly downloaded in bulk via FTP (ftp://ftp.ensemblgenomes.org).
Ensembl Genomes data is also made available through a series of data warehouses, optimized around common (gene and SNP-centric) queries, using the BioMart data warehousing system (4,5). The BioMart framework provides a series of interfaces, including web-based query building tools, for each of the Ensembl Genomes domains (e.g. at http://plants.ensembl.org/biomart/martview) and a variety of other interfaces for interactive and programmatic access.
Ensembl Genomes is released four to five times a year, in synchrony with releases of Ensembl, utilizing the same software as the corresponding Ensembl release. The overall suite of Ensembl Genomes interfaces mirrors the interfaces provided for vertebrate genomes in Ensembl, and allows users access to genomic data from across the tree of life in a consistent manner.
COLLABORATION AND ANNOTATION
Over 3000 genomes have been sequenced, annotated and deposited in the archives maintained by the International Nucleotide Sequence Database Collaboration. While these genomes are mostly bacterial, the number of eukaryotic species sequenced already exceeds 200; and increasing quantities of data are available for most of these species, with resequencing of large populations of individuals (to identify polymorphism) now common in all parts of the taxonomy. These developments have created a need for sophisticated systems [e.g. Ensembl, the Generic Model Organism Database toolset (http://gmod.org), the UCSC genome browser (6)] for the management of genome-scale data; but the growth in data volumes (the nucleotide archives are presently doubling in size every 9 months) naturally raises questions of future scalability. In the case of annotated genomes, such concerns are focused less on absolute data volume (as an annotated genome sequence is itself a compacted, interpreted form of the raw data present in the archives), but on the human resources needed to integrate and interpret the raw data correctly. Pipelines for constructing Ensembl databases are highly automated, but owing to differences in the biology and available data for each species, the production of high quality annotation still depends on a measure of manual intervention (for parameterization, quality control and biological validation). Expert groups focused on particular organisms exist in many domains, but sometimes lack the resources to develop infrastructure to support the increasing variety of available data types. Moreover, cross-species analysis may be difficult when data is distributed across different sites, each with their own technical implementations.
In Ensembl Genomes, we address these problems by collaborating with community-based groups maintaining specialized resources, to assist in the process of genome annotation and to provide a permanent integrative portal for data from many species. The form of these collaborations depends on the mandate and expertise of the collaborating group, and the fit of their data to the Ensembl toolset. In domains where a well-established resource (such as a model organism database) is already maintaining primary annotation, this information is integrated into the Ensembl data structure, and supplemented with additional high-value data sets where available. In many cases, Ensembl Genomes actively participates in these community-based initiatives, directly contributing to the production of reference annotation. In these partnerships, each project's own portal remains the primary point of access for researchers working within the relevant domain, and will often display a wide range of data types; while Ensembl Genomes places the project's genome-centric data in its broader context. We believe that this model for collaboration—involving specialized resources, which take custodianship of collections of closely related genomes and which are grounded in their own communities; the re-use of technology and expertise; and the provision of a pan-taxonomic integrating portal—is a viable response to increasing data growth.
In the plant domain, we work with Gramene (http://www.gramene.org) (7), a peer resource (also utilizing Ensembl technology) to maintain a common resource for plant genomics in Europe and USA. We have also recently established a new collaborative network, transPLANT, to develop a European-wide infrastructure for genome scale data in plants. We are part of the VectorBase (http://www.vectorbase.org) consortium (8), a NIH NIAID resource for the genomes of invertebrate pathogens of human diseases. We have recently joined WormBase (http://www.wormbase.org) (9), which maintains resources for nematode genomes, especially the model species Caenorhabditis elegans. Two recent collaborations have established PomBase (http://www.pombase.org), a new model organism database focused on the fission yeast Schizosaccharomyces pombe; and PhytoPath (http://www.phytopathdb.org), a resource for plant pathogens, with a focus on fungi and oomycetes. These two projects are utilizing Ensembl software for data visualization. Additionally, we have been working with the Central Aspergillus Data Respoistory, CADRE (http://www.cadre-genomes.org.uk) (10) on genomes of the genus Aspergillus; and with the Broad Institute and the U.S. Agricultural Research Service on the genome of the wheat stem rust pathogen, Puccinia graminis. We are always open to new collaborations to extend the range of species covered, and to analyse and integrate the output of specific scientific projects.
NEW CONTENT: BREADTH AND DEPTH
With so much data available, Ensembl Genomes has to prioritize data for incorporation. Priorities are firstly, data relevant to our specific collaborations; secondly, data from other major experimental species and thirdly, data from other species that provide local or remote evolutionary context for the priority species, and which are used to strengthen the comparative analysis provided in the site. For the first category of genomes, we actively work with our collaborators to produce the primary, community-recognized annotation. For the second category, we supplement the reference annotation (often maintained by model organism databases or other similar resources) with additional high-value data sets. For several species in these two categories, we have built variation databases, which store genotypes, loci and phenotypes from large-scale genome-wide array-based and resequencing studies; the data is available through a suite of specialized graphical views and a SNP-centric BioMart. Variation data is sourced from dbSNP (11) where available, or otherwise directly from the data producers. For the third category of genomes, annotation is generally incorporated from the original submitters with only limited enhancement (e.g. the annotation of non-coding genes, if absent in the original submission).
At the time of writing, there have been seven releases of Ensembl Genomes since the previous report was published in this journal. The current release is release 10, made public in July 2011. In this time, there has been a significant increase in the content of all five Ensembl Genomes sites. A complete list of genome species in the databases, and the source of their annotation, is provided in Supplementary Table S1.
Metazoa
New species added include the body louse Pediculus humanus (from VectorBase), Pristionchus pacificus (from WormBase), the pea aphid Acyrthosiphon pisum, the honey bee Apis mellifera, the sea urchin Strongylocentrotus purpuratus, the blood fluke Schistosoma mansoni, the sea anemone Nematostella vectensi and the basal metazoan Trichoplax adhaerens.
A variation database has been added for Drosophila melanogaster, using data from the Drosophila Population Genomics Project. Gene builds have been updated for various species, including Aedes aegypti and Anopheles gambiae, and mappings to an increased range of microarray probes added for A. gambiae and C. elegans. DNA-centric comparative analysis databases have been added for drosophilids, worms and mosquitoes.
Plants
New species added include maize Zea mays, African rice Oryza glaberrima and the moss Physcomitrella patens. Updated databases have been produced for Arabidopsis thaliana [based on the TAIR10 release (12)], Vitis vinifera, poplar Populus trichocarpa and rice O. sativa indica.
The variation database for A. thaliana has been regularly updated, accommodating the latest data made available from a number of major genotyping initiatives in this species. The variation resource for O. sativa japonica has also been enhanced. Mappings to an increased range of microarray probes have been added for A. thaliana and both cultivar groups (indica and japonica) of rice.
Fungi
New species added include the bread mould Neurospora crassa, the wheat stem rust parasite P. graminis f. sp. tritici, and six other plant pathogens: Fusarium oxysporum, Gibberella moniliformis, G. zeae, Ustilago maydis, Nectria haematococca and P. triticina.
Updated databases have been generated for Aspergillus fumigatus, A. nidulans and S. pombe. A variation database has been produced for G. zeae.
Protists
New species added include the slime mould Dictyostelium discoideum [using data from dictyBase (13)], the diatoms Phaeodactylum tricornutum and Thalassiosira pseudonana, two additional malarial parasites, Plasmodium chaubaudi and P. berghei, four oomycete plant pathogens: Phytophthora infestans, P. ramorum, P. sojae and Pythium ultimum; and the kinetoplastid parasites Leishmania major and Trypanosoma brucei. Updated microarray probe mappings have been generated for P. falciparum. A variation database has been constructed for P. falciparum, and a DNA-centric comparative analysis databases has been added for Phytophthora species.
Bacteria
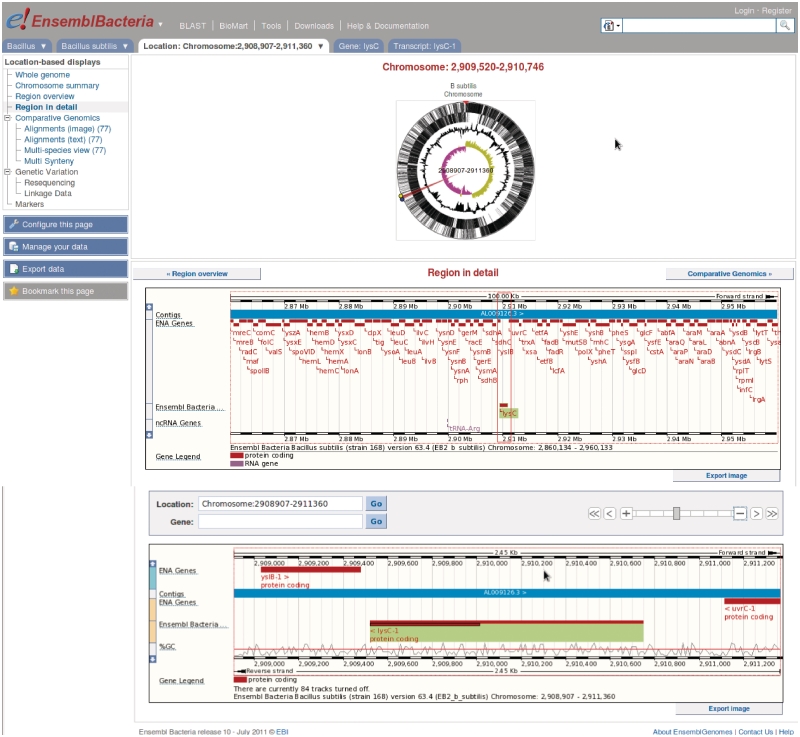
Ensembl Bacteria is organized around clades of closely related bacterial species. Since the launch of Ensembl Bacteria, we have added new databases representing the Boreila, Buchnera and Wollbachia genera. These genera have been prioritized because their members have symbiotic relationships with insect species already present in Ensembl Metazoa, and are in some cases the causative agents of human or animal disease. The total number of individual genomes represented in Ensembl Bacteria has risen from 114 at launch to a current total of 249; the web browser has been revised to display circular chromosomes (which are normal in bacteria) in an appropriate way (Figure 1); and the data structure revised to enable the correct modelling of polycistronic transcripts.
Figure 1.
Visualizing bacterial genomes in Ensembl Genomes. The Ensembl browser has been customized in various ways to support the visualization of bacterial genes and genomes. The panel shows the navigation tool for circular genomes (top), and the representation of alternate translations within one transcript (bottom).
The source of the gene annotations displayed in Ensembl Bacteria is mostly depositions to the public nucleotide sequence archives (14). However, the total number of sequenced bacterial genomes deposited here is now ∼3000. We are developing a new gateway to provide comprehensive availability of these (and future) genomes through an Ensembl-like interface, while continuing to feature specific clades (where there is sufficient data/community interest to drive improvements or additions to the submitted annotation) in the Ensembl Bacteria portal.
COMPARATIVE ANALYSIS
Extensive comparative analyses are performed between the sequences in Ensembl Genomes. Analyses include pairwise alignments between DNA sequences, using the tools LASTZ (15) and (for more diverged genomes) translated BLAT (16), combined with the use of the UCSC chain/net algorithm (17); and multiple alignments using Enredo and Pecan (18). Protein alignments are used to reconstruct evolutionary trees for related genes using the Ensembl Compara Gene Trees pipeline (19). These are run for each individual domain and additionally, for a representative selection of 47 species chosen from the entire taxonomic space, which identifies widely conserved families and deep homologies between different evolutionary branches.
VARIATION RESOURCES
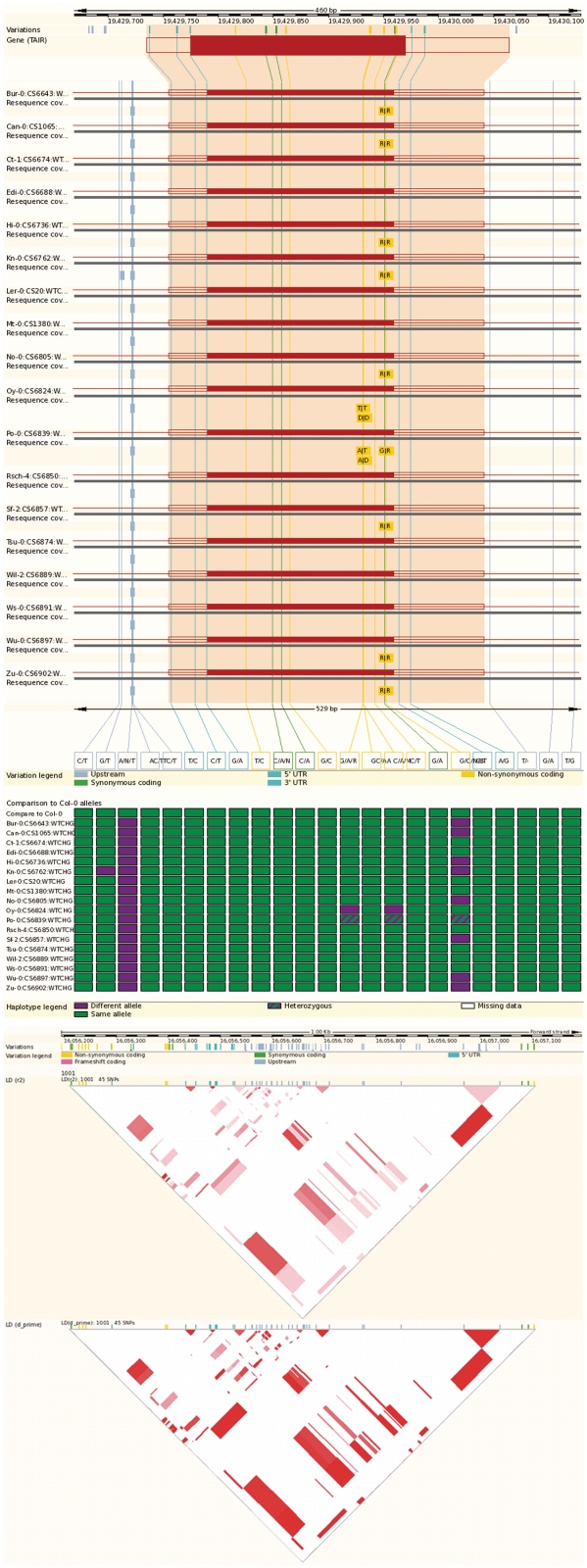
Identification of polymorphism, on a genome wide scale in both natural populations and known breeding stocks, is increasingly common in many species. In response to this, Ensembl Genomes has been developing resources to store and provide access to such data in the context of reference genome sequences. We now have variation resources for nine species, including two cultivars of rice, grape, the malarial vector A. gambiae and the malarial parasite P. falciparum, the phytopathogen G. zeae, Saccharomyces cerevisiae [using data from the Saccharomyces Genome Resequencing Project (20)] and D. melanogaster, where we represent the data generated in the Drosophila Population Genome Project (http://www.dpgp.org). The most complicated data set currently represented is that for A. thaliana, where we have integrated array-based genotypes with data from re-sequencing approaches from the 1001 Genomes project (http://www.1001genomes.org) and other studies (21,22). Some of these data sets are very large: the Arabidopsis variation database already comprises over 14 million variants from over 1600 sequenced accessions, with a total of over 350 million individual (non-reference) locus genotypes reported. Even larger data sets are likely to be generated soon for a number of species, in particular for crop plants. Ensembl Genomes facilitates access to these data through graphical interfaces supporting the visualization of variants in the context of their associated genomic location, gene, transcript or protein sequence; the functional consequences of each variant, and linkage disequilibrium between variant loci (Figure 2). Additionally, since 2010, variant-centric BioMarts have been provided, allowing the download of large subsets of data matching user-specified criteria.
Figure 2.
Visualizing genome-scale variation in Ensembl Genomes. There are various views allowing users to explore identified genomic variation in the context of reference annotation. The figure shows (using data from A. thaliana) the ‘transcript comparison page’ (top panel), which displays the location and functional consequences of each variant in the context of its location within a transcript, and the distribution of alleles among individuals (wild type, homozygous variant or heterozygous) using a colour coded tabular display; and the linkage disequilibrium view (lower panel).
UPLOAD YOUR DATA
A major improvement to the Ensembl Genomes site has been the development of more ways for users to upload their own data to the site. Users can operate a server meeting the specifications of the Distributed Annotation System (23), which allows them to make their data available to all users of the site and additionally, to other websites that (like Ensembl) function as DAS clients. Users can also upload their data to a private area of the Ensembl database; or direct their browsers to load data dynamically from local files available over HTTP or FTP. The range of supported file formats has increased and not only includes traditional formats for genome annotation (e.g. GFF) but also formats including BAM, BigWig and VCF, typically used to store the large data associated with next generation sequencing technology. This facility enables users to see genome-transcriptome alignments and variant calls (based on their own data) in the context of the reference annotation; and is likely to grow in importance with the continued adoption of high throughput sequencing as a routine laboratory technique.
PERSPECTIVES AND PRIORITIES
The future development of Ensembl Genomes is likely to continue to be shaped by the increasing quantities of genome sequencing being undertaken. This has positive consequences: more genomes of interest are sequenced, the quality of annotation is improved (e.g. RNA-seq is already beginning to provide much greater coverage of the gene space than traditional EST libraries, enabling the production of better-supported gene models and the identification of new variants), and a large amount of knowledge about sequence variation can be generated. Data sets will continue to be prioritized according to community demand, concentrating on the species of greatest scientific interest and those that provide the most useful scientific context.
To consider some more specific challenges, it is likely that the next 2 years will see a draft annotated assembly of the wheat genome, which is both large (16 GB) and hexaploid. Polyploidy defines a new logical framework for the representation of variation and evolution, and there will be a need to update schemas and interfaces accordingly. Another challenge will come from incorporating data from large-scale projects aimed at deriving sequence from large numbers of closely related (but distinct) species (e.g. the 15 Anopheles genomes currently being annotated by VectorBase http://www.vectorbase.org/sections/Other/addtl_org_includes/Anopheles_Genomes_4Aug08.pdf). In contrast to intra-species resequencing projects, it will often be desired to annotate these genomes (instead of just calling variants), but unlike a conventional single species sequencing project, a logical strategy might be to combine the use of inference from a single well-annotated reference combined with standardized experimental techniques (e.g. RNA-seq for transcriptome determination) applied across all species in the project. As greater numbers of closely related species are sequenced, the data model already used for Ensembl Bacteria, in which related genomes are grouped in collections, may become increasingly applicable to other domains. However, the historical model organisms are likely to remain the focus of most of the experimentally-validated functional information from which their annotation will be derived. While it does not seem plausible to annotate every genome to the standard of the existing model species, the challenge is to find scalable ways of enabling the reference annotation to inform the biology of the widest possible species set.
Another challenge will come from structural variation, which is widespread even within species, but which is poorly represented by most genome browsers. Possible reasons for this include the shortage of reliable data (as the short read technologies that have revolutionized sequencing often fail to provide long-range information); but also, the convenience and power of the paradigm of the linear genome as a template for functional annotation, a paradigm that most browsers adhere to. High levels of structural rearrangement can only properly be represented using graphical paradigms, and we are accordingly developing new interfaces to visualize linear genomes in the context of structural diversity.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Table S1.
FUNDING
The United Kingdom Biosciences and Biotechnology Research Council (grants BB/I00I0077/1, BB/H531519/1, BB/F19793/1); Wellcome Trust (grant 090548/B/09/Z); Bill and Melinda Gates Foundation (grant OPPGD1491); framework 7 programme of the European Union (contracts 226073, Sling; 228421, INFRAVEC and 284496, transPLANT). Funding for open access charge: Institutional budget (European Molecular Biology Laboratory).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We would also like to acknowledge the contributions of all our collaborators; and of all colleagues working on the Ensembl project at the EBI and the Wellcome Trust Sanger Institute.
REFERENCES
- 1.Flicek P, Amode MR, Barrell D, Beal K, Brent S, Chen Y, Clapham P, Coates G, Fairley S, Fitzgerald S, et al. Ensembl 2011. Nucleic Acids Res. 2010;39:D800–D806. doi: 10.1093/nar/gkq1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.The UniProt Consortium. The Universal Protein Resource (UniProt) in 2010. Nucleic Acids Res. 2009;38:D142–D148. doi: 10.1093/nar/gkp846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Quevillon E, Silventoinen V, Pillai S, Harte N, Mulder N, Apweiler R, Lopez R. InterProScan: protein domains identifier. Nucleic Acids Res. 2005;33:W116–W120. doi: 10.1093/nar/gki442. doi:10.1093/nar/gki442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kinsella RJ, Kähäri A, Haider S, Zamora J, Proctor G, Spudich G, Almeida-King J, Staines D, Derwent P, Kerhornou A, et al. Ensembl BioMarts: a hub for data retrieval across taxonomic space. Database. 2011;2011 doi: 10.1093/database/bar030. doi:10.1093/database/bar030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Smedley D, Haider S, Ballester B, Holland R, London D, Thorisson G, Kasprzyk A. BioMart–biological queries made easy. BMC Genomics. 2009;10:22. doi: 10.1186/1471-2164-10-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fujita PA, Rhead B, Zweig AS, Hinrichs AS, Karolchik D, Cline MS, Goldman M, Barber GP, Clawson H, Coelho A, et al. The UCSC Genome Browser database: update 2011. Nucleic Acids Res. 2011;39:D876–D882. doi: 10.1093/nar/gkq963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Youens-Clark K, Buckler E, Casstevens T, Chen C, Declerck G, Derwent P, Dharmawardhana P, Jaiswal P, Kersey P, Karthikeyan AS, et al. Gramene database in 2010: updates and extensions. Nucleic Acids Res. 2011;39:D1085–D1094. doi: 10.1093/nar/gkq1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lawson D, Arensburger P, Atkinson P, Besansky NJ, Bruggner RV, Butler R, Campbell KS, Christophides GK, Christley S, Dialynas E, et al. VectorBase: a data resource for invertebrate vector genomics. Nucleic Acids Res. 2009;37:D583–D587. doi: 10.1093/nar/gkn857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harris TW, Antoshechkin I, Bieri T, Blasiar D, Chan J, Chen WJ, De La Cruz N, Davis P, Duesbury M, Fang R, et al. WormBase: a comprehensive resource for nematode research. Nucleic Acids Res. 2009;38:D463–D467. doi: 10.1093/nar/gkp952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mabey JE. CADRE: the Central Aspergillus Data REpository. Nucleic Acids Res. 2004;32:401D–405D. doi: 10.1093/nar/gkh009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sherry ST, Ward MH, Kholodov M, Baker J, Phan L, Smigielski EM, Sirotkin K. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Swarbreck D, Wilks C, Lamesch P, Berardini TZ, Garcia-Hernandez M, Foerster H, Li D, Meyer T, Muller R, Ploetz L, et al. The Arabidopsis Information Resource (TAIR): gene structure and function annotation. Nucleic Acids Res. 2007;36:D1009–D1014. doi: 10.1093/nar/gkm965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gaudet P, Fey P, Basu S, Bushmanova YA, Dodson R, Sheppard KA, Just EM, Kibbe WA, Chisholm RL. dictyBase update 2011: web 2.0 functionality and the initial steps towards a genome portal for the Amoebozoa. Nucleic Acids Res. 2011;39:D620–D624. doi: 10.1093/nar/gkq1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cochrane G, Karsch-Mizrachi I, Nakamura Y. The International Nucleotide Sequence Database Collaboration. Nucleic Acids Res. 2011;39:D15–D18. doi: 10.1093/nar/gkq1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harris RS. Improved pairwise alignment of genomic DNA. 2007 Ph.D. Thesis. The Pennsylvania State University. [Google Scholar]
- 16.Kent WJ. BLAT–the BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kent WJ, Baertsch R, Hinrichs A, Miller W, Haussler D. Evolution's cauldron: duplication, deletion, and rearrangement in the mouse and human genomes. Proc. Natl Acad. Sci. USA. 2003;100:11484–11489. doi: 10.1073/pnas.1932072100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Paten B, Herrero J, Beal K, Fitzgerald S, Birney E. Enredo and Pecan: genome-wide mammalian consistency-based multiple alignment with paralogs. Genome Res. 2008;18:1814–1828. doi: 10.1101/gr.076554.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vilella AJ, Severin J, Ureta-Vidal A, Heng L, Durbin R, Birney E. EnsemblCompara GeneTrees: complete, duplication-aware phylogenetic trees in vertebrates. Genome Res. 2009;19:327–335. doi: 10.1101/gr.073585.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liti G, Carter DM, Moses AM, Warringer J, Parts L, James SA, Davey RP, Roberts IN, Burt A, Koufopanou V, et al. Population genomics of domestic and wild yeasts. Nature. 2009;458:337–341. doi: 10.1038/nature07743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gan X, Stegle O, Behr J, Steffen JG, Drewe P, Hildebrand KL, Lyngsoe R, Schultheiss SJ, Osborne EJ, Sreedharan VT, et al. Multiple reference genomes and transcriptomes for Arabidopsis thaliana. Nature. 2011 doi: 10.1038/nature10414. 10.1038/nature10414. http://dx.doi.org/10.1038/nature10414 (13 September 2011, date last accessed) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Atwell S, Huang YS, Vilhjálmsson BJ, Willems G, Horton M, Li Y, Meng D, Platt A, Tarone AM, Hu TT, et al. Genome-wide association study of 107 phenotypes in Arabidopsis thaliana inbred lines. Nature. 2010;465:627–631. doi: 10.1038/nature08800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jenkinson AM, Albrecht M, Birney E, Blankenburg H, Down T, Finn RD, Hermjakob H, Hubbard TJP, Jimenez RC, Jones P, et al. Integrating biological data–the Distributed Annotation System. BMC Bioinformatics. 2008;9(Suppl. 8):S3. doi: 10.1186/1471-2105-9-S8-S3. [DOI] [PMC free article] [PubMed] [Google Scholar]