Abstract
Integrated Pathway Resources, Analysis and Visualization System (iPAVS) is an integrated biological pathway database designed to support pathway discovery in the fields of proteomics, transcriptomics, metabolomics and systems biology. The key goal of IPAVS is to provide biologists access to expert-curated pathways from experimental data belonging to specific biological contexts related to cell types, tissues, organs and diseases. IPAVS currently integrates over 500 human pathways (consisting of 24 574 interactions) that include metabolic-, signaling- and disease-related pathways, drug–action pathways and several large process maps collated from other pathway resources. IPAVS web interface allows biologists to browse and search pathway resources and provides tools for data import, management, visualization and analysis to support the interpretation of biological data in light of cellular processes. Systems Biology Graphical Notations (SBGN) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway notations are used for the visual display of pathway information. The integrated datasets in IPAVS are made available in several standard data formats that can be downloaded. IPAVS is available at: http://ipavs.cidms.org.
INTRODUCTION
In the past decade, there has been accumulation of large mass of biological data by the use of high-throughput omics technologies (e.g. genomics, transcriptomics, proteomics and metabolomics). Biological pathways can represent complex processes at molecular level and can be a valuable aid for computational and experimental research utilizing the omics data (1). Biologists can use pathway databases equipped with easy-to-use analytical and visualization tools to garner insight about their experiments (e.g. genome wide association studies, next generation genome sequencing projects and molecular profiling data), digest large amounts of information and generate hypotheses.
There are several manually curated publically available pathway resources, including PANTHER (2), Reactome (3), KEGG (4), MetaCyc (5), WikiPathways (6), PharmGKB (7), SMPDB (8), PID (9) and large process maps frequently published by the Systems Biology Institute (SBI) (10,11) and deposited in Payao (12). Several companies provide open-access to curated pathway databases such as Qiagen's GeneGlobe Pathway Central (https://www.qiagen.com/geneglobe/pathways.aspx), BioCarta pathways (http://www.biocarta.com),) and Ambion’s Pathway Atlas (http://www.ambion.com/tools/DARKSITE/pathway/all_pathway_list.php). Additionally a number of commercial pathway databases such as GeneGo's Pathway Maps (http://www.genego.com/mapbrowse.php) and Ingenuity Pathway Analysis tool (http://www.ingenuity.com/) are also available.
Integrated Pathway Resources, Analysis and Visualization System (IPAVS) is a freely available, interactive and integrated pathway database which is designed to address the needs of bench biologists, computational biologists and physicians. It offers biologists a single point of access to several manually curated pathway resources, in addition to its own expert-curated pathways that are in standard format.
UNIQUE FEATURES AND COMPARISONS OF PATHWAY DATABASES
Most of the aforementioned databases including IPAVS consist of a mix of metabolic, signaling and disease pathways. Some databases emphasize a particular type of pathways such as drug pathways (PharmaGKB), metabolic pathways (SMPDB, MetaCyc and Reactome) or signaling pathways (PID). Many databases have their contents curated by a team of experts (e.g. PANTHER, Reactome, KEGG, MetaCyc, PharmaGKB, SMPDB) and provide access to only their curated pathways. Databases such as Payao and Wikpathways are collaborative web service platforms which mainly depend upon the community to provide annotations and curated pathways. Although overall quality of information and coverage of most of the databases mentioned are quite impressive, there is still vast room for improvement. Most pathways in some of the above-mentioned databases are generic and have not been curated in any specific biological context. However, we believe that building pathways in specific contexts will allow gathering of more unique information and help prevent redundancy. To this end, pathways in IPAVS are curated in specific biological themes or contexts, such as type of cell, tissue, or organ, phenotypes and diseases, toxicological exposure, and various perturbed conditions, that are not covered or are scantly covered in other databases.
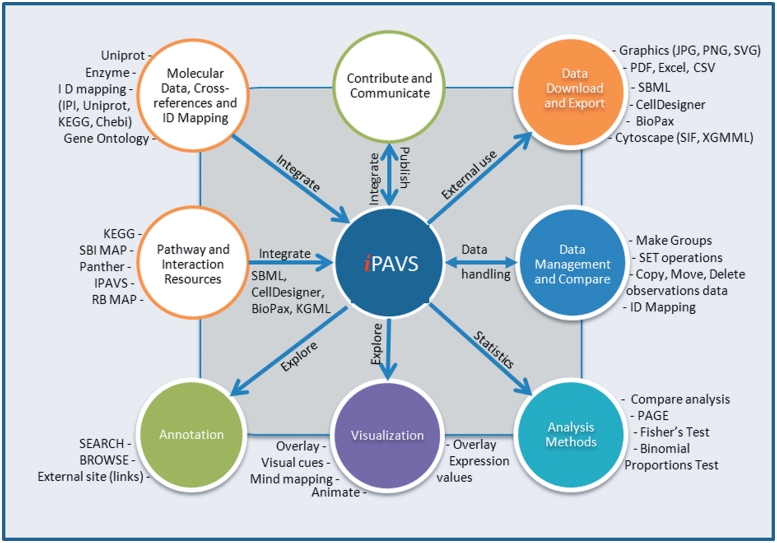
Most pathway databases provide simple searches and browsing of pathway information and few such as Reactome, MetaCyc and KEGG support mapping and visualization of the gene, protein expression and/or metabolite data onto pathway diagrams. Databases like PathwayCommons (13), PID and Reactome support analysis tools and statistical algorithms for conducting systematic pathway enrichment analysis. ConsensusPathDB (14) and PathwayCommons (13) collate data from several sources and provide web services enabling biologists to browse and search comprehensive collections of pathway data from multiple sources and carry statistical analysis with integrated data. However, there are very few databases like PID which provide their own curated data and also integrate information from multiple databases. IPAVS provides human signaling and metabolic pathways curated in a specific biological context and integrates five pathway resources (Table 1). In addition, IPAVS provides several tools to support visualization and analysis for interpretation of user-specified gene or protein expression data and metabolite data (Figure 1). All data in IPAVS is freely available without any restriction, and all datasets can be downloaded.
Table 1.
Summary of all data sources
| Completely curated | Imported (with partial curation) | Automatically imported | |||
|---|---|---|---|---|---|
| Datasets | IPAVS | Panther (2) | SBI-MAPs (6,7) | RB-Maps (5) | KEGG (Human) (4) |
| Pathways | 60 | 165 | 7 | 17 | 234 |
| Pathway types | Signaling, metabolic, GNR, disease map (e.g. cancer, hypertrophy, heart failure, aciduria, hypermethoninemia, etc.) | Signaling, metabolic, disease map | Signaling, metabolic | Signaling | Signaling, metabolic, disease MAP, organismal, GNR |
| Pathway context | Survival, development, adhesion, cardioprotection, cell growth and death, stress induced, EC coupling, stretch activated, cell and tissue specific and others | Few pathways of diseases and physiology | Cell specific | Digestive, endocrine, excretory, nervous, immune, developmental, cell growth and death, membrane transport and others | |
| Interaction | 3115 | 5043 | 4275 | 689 | 11 452 |
| Proteins/gene/RNA | 910 (∼30% only in IPAVSa) | 1758 | 1110 | 81 | 4315 |
| Protein modifications | 380 | 736 | 1235 | 298 | 590 |
| Small molecule | 386 (∼20% only in IPAVSa) | 749 | 231 | 2700 | |
| Complexes | 363 | 558 | 333 | 62 | 669 |
| Phenotype | 117 (∼80% only in IPAVSa) | 109 | 24 | 0 | (Annotated as image) Not available for computation |
| PMID (level annotated) | 1688 (P, I and few C) | 1953 (P) | 640 (P and I) | 141(P and I) | 2105 (P) |
aSee Supplementary File S1 for the complete list.
GNR = gene regulatory network; P = pathway; I = interaction; C = complex.
Figure 1.
Overview of IPAVS pathway resources and web application features.
DATA
The IPAVS data model was formulated to import and integrate datasets that are available in two largely used standards—BioPax (15) and SBML (with CellDesigner extensions) (16). Pathways in IPAVS include biochemical reactions, complex assembly, transport, catalysis and inhibitory events and physical interactions involving molecules (proteins, genes, RNA, antisense RNA, compounds/small molecules and ions) and supramolecular complexes. Large maps interlink several pathways in a specific biological context (tissue, time, perturbation, disease/phenotype, physiology). Additionally, all IPAVS curated pathways and maps include information on relevant organs, tissues, organelles, subcellular location of molecules, post-translational modifications, activity states of molecules, descriptions providing an overview of the pathway and supporting experimental evidence for the pathway and each of its interactions.
DATA CURATION, IMPORTING DATA AND DATABASE CONTENT INFORMATION
One of the goals of IPAVS is to provide a manually curated pathway resource. IPAVS has adopted an incremental and iterative curation work process. The curation steps involve identifying and organizing the required literature content (primary journal articles and review papers). The relevant information is extracted, verified and then assembled into prototype pathway maps using CellDesigner (software for pathway diagram editors) (16), which is then gradually refined and annotated with all the curated information including associating every molecule with an standard controlled identifiers, evidence information for pathways and interactions, and description providing an overview of pathways to obtain an accurate, information rich pathway model.
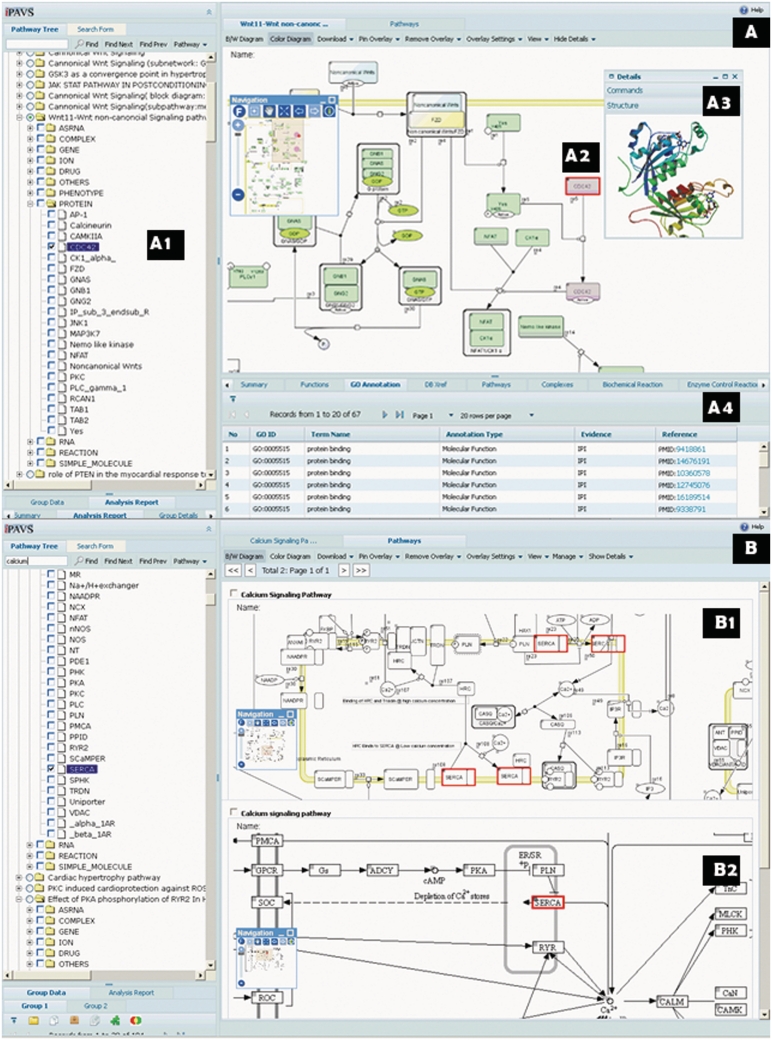
IPAVS complements existing resources by providing pathways that are curated in specific biological themes or contexts. For example, calcium signaling pathways from IPAVS and KEGG are compared in Figure 2B. The pathway curated by IPAVS [Figure 2B(1)] represents data obtained from cardiomyocytes and has numerous molecules (34 entities), interactions and supporting annotations (154 PubMed entries) that are not present in the pathway curated by KEGG [Figure 2B(2)]. This is because many of the molecules regulating calcium homeostasis in cardiomyocytes have tissue-specific expression and are not expressed in other tissues. Therefore, pathways that are designed to be very generic and are not curated in a particular context (e.g. cell, tissue or organ type), such as the one from KEGG, could be missing information that can be found in IPAVS. Differences can also be noticed at the levels of intent and extent of pathway coverage. Most of the existing generic pathway databases like KEGG, PANTHER and Reactome have very few pathways related to disease, drug or other aforementioned contexts. While KEGG provides drug pathways focused on drug development or drug similarity, IPAVS’ drug pathways often capture drug's action or mechanism. Therefore, IPAVS not only has enhanced information in regard to description of existing pathways in a particular context, but also has additional content that is not normally found in other pathway databases. The context-curated pathways are more relevant to biologists as they can provide them with information specific to their needs. This is evident from the high number of biologists who refer to the website (http://cidms.org/pathways/er_stress/index.html) that hosts Endoplasmic Reticulum Stress Response interactive pathway (17). With the availability of information-rich pathway sets, well-known pathway analysis methods could be adjusted for the framework of different tissue types, pathologies and numerous other biological contexts, thus allowing the accurate deduction of biological meaning from the data (18).
Figure 2.
(A) Single pathway view showing the non-canonical Wnt-signaling pathway. Protein CDC42 (highlighted) was clicked on the diagram [(A2) red rectangle], triggering the highlighting of the corresponding node on the tree (A1), the coloring of all its instances in the diagram (A2 red rectangle) and also the opening of the ‘Details’ panel (showing GO annotations) at the bottom (A4). Additionally, a context-popup-window displays the structure of CDC42 (A3). (B) Multi-pathway view showing the calcium signaling pathway, with the view zoomed in to the Sarcoplasmic region. The pathway from IPAVS curated in the context of cardiac tissue (top) is compared with corresponding pathway from KEGG. The IPAVS pathway shows many more interactions specific to cardiac tissue regulating calcium homeostasis.
IPAVS integrates data from five pathway sources (Table 1). Several manually curated resources of large process maps (10,11) that are superior in terms of reliability and detail, and can aid in the generation of biologically meaningful hypotheses are available. Unfortunately, until now this information has not been integrated into any existing public databases, making it difficult for researchers to access it. We have collected, verified and manually annotated the missing information before some of these pathways could be integrated into IPAVS. Also, IPAVS has been designed to automatically integrate data from other pathway database like PANTHER database (2) and KEGG (4) using custom written loaders and converters.
DIAGRAM NOTATION
SBGN is a community accepted standard of visual languages that helps biologists communicate complex pathways without any ambiguity. The IPAVS pathway diagrams mostly use SBGN (19) and KGML notation for KEGG pathways. Although KGML pathways were successfully converted into SBGN notation, for the sake of clarity, KEGG pathway diagrams are still used in their original format instead of being automatically laid out (which could produce messy outputs for large pathway diagrams).
USING THE IPAVS WEB APPLICATION
Browse, search and visualize pathway information
The IPAVS user interface (UI) is designed to allow users to browse and search pathway information across multiple pathway resources. The UI has four main panels that allow quick and easy access to the tools needed to explore pathway information. User can use ‘Pathway Browser’ panel (left side of UI) to quickly click down the hierarchy of pathway information and locate molecules or interactions participating in the pathways. Clicking on a pathway in the ‘Browser’ displays the corresponding pathway diagrams in the ‘Visualization’ panel. Users can zoom, pan and navigate different regions in the pathway diagram. Researchers can interact with one pathway or multiple pathways as a group. In group view, pathways or pathway overlaid with analysis data can be compared and contrasted (Figure 2B). The contextual details of pathways and any of its individual components can also be viewed in ‘Details Panel’.
IPAVS supports a full search feature that is implemented using the Apache–Lucene text indexing and search engine (http://lucene.apache.org/), which allows keywords, quoted phrases, wild cards and Boolean queries. Users can search molecules, interactions and pathways by entering a name or accession number (e.g. Uniprot, Chebi and PMID) or some associated term(s). By clicking on links provided with every record in the result, its relevant details can be viewed. Furthermore, users can set filters to customize the search query, restricting it to specific organisms, databases or particular datasets.
Data upload, data management and comparison
IPAVS allows for the investigation of a variety of omics data in the context of cellular pathways. Users can upload data using the upload wizard. IPAVS supports a wide variety of gene, protein and metabolite identifiers, allowing user data to be more completely connected to the pathways in IPAVS. Similar to how biologists design and organize their experiments in groups, in IPAVS the uploaded data can be organized into logical groups. Furthermore, users can employ data management tools, allowing copy, move and delete operations on the group records to enable disparate datasets to be combined in some biological context. Such groups (contextual subsets) created for particular genes of interest can help users to track the gene and its context during the analysis. If a user is interested in comparing groups, he can use the ‘Comparison’ tool that provides SET operations that can find the intersections and differences among the compared groups.
Pathway and expression analysis supported with visualization
IPAVS currently implements three analysis algorithms following two approaches: (i) Fishers Exact test and Binomial proportions test for statistically testing the significance of the overlaps between user data and pathways (20) and (ii) parametric analysis of gene set enrichment (PAGE) to measure and compare whether a pathway shows a consistent trend towards stronger phenotypes (21). After uploading the data, the ‘Analysis Wizard’ can be used for executing analysis tasks. Users can customize various parameters of analysis including setting filters to include only a specific set of pathways meeting certain criteria or a biological context. The analytical capability of IPAVS is intricately integrated with a broad range of visualizations that help to generate meaningful insights. The quantitative data (e.g. gene expression) of molecules can be overlaid as color, shapes, embedded small charts (line or bar) and heat maps on the pathways.
Data download and export
IPAVS allows the export of pathways to various graphical and machine-readable standard file formats (SBML, BioPAX, XGMML and CD) and convenient file formats (SIF, tab-delimited, CSV files) individually, in batches or all at once (bulk download). Users can also save the entire pathway map or specific zoomed regions along with visual annotations (charts or heat maps of expression data) that were overlaid during the pathway exploration and analysis.
COMMUNITY CONTRIBUTIONS
Currently, the community can contribute in two ways. First, experts can curate new pathways or even download pathways from IPAVS and modify/update them remotely using the CellDesigner tool, and submit them by email (support@cidms.org). Second, users can submit functional annotation as concise phrases describing an entity or events in the pathway along with evidence (complete citation or PMID) using web form. The information will be verified by the IPAVS team and then made available to the public. Support for curation training and reviewing of pathways is available by request from the IPAVS team.
FUTURE PERSPECTIVES
IPAVS is an ongoing project. We are continuously adding five to six pathways every month and constantly revising existing pathways. At present the data in IPAVS has not been merged (i.e. if two sources describe the same pathway, IPAVS does not create a single unified pathway), however we will work towards this in the near future. We have also planned several enhancements for integrating additional pathway (e.g. Reactome) and interaction [e.g. HPRD (22), MINT (23)] resources including non-human data, visualization (‘on the fly’ rendering of pathway maps using Cytoscape Web (http://cytoscapeweb.cytoscape.org/), analysis [topology based enrichment analysis (24)] and data management capabilities. Please see the online wish list (http://ipavs.cidms.org/wish-list) for details.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary File S1.
FUNDING
Funding for open access charge: Korea MEST NRF Grant (2011-0002144) and GIST Systems Biology Infrastructure Establishment Grant (2011).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors would like to thank S.K.A. Siva and all developers of open-source for their contributions to software we use, without which our task would have been impossible. The authors appreciate the dedicated efforts of data curators and institutions for making the curated information freely available to the community.
REFERENCES
- 1.Kelder T, Conklin BR, Evelo CT, Pico AR. Finding the right questions: exploratory pathway analysis to enhance biological discovery in large datasets. PLoS Biol. 2010;8:e1000472. doi: 10.1371/journal.pbio.1000472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mi H, Dong Q, Muruganujan A, Gaudet P, Lewis S, Thomas PD. PANTHER version 7: improved phylogenetic trees, orthologs and collaboration with the Gene Ontology Consortium. Nucleic Acids Res. 2010;38:D204–D210. doi: 10.1093/nar/gkp1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Croft D, O'Kelly G, Wu G, Haw R, Gillespie M, Matthews L, Caudy M, Garapati P, Gopinath G, Jassal B, et al. Reactome: a database of reactions, pathways and biological processes. Nucleic Acids Res. 2011;39:D691–D697. doi: 10.1093/nar/gkq1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kanehisa M, Goto S, Furumichi M, Tanabe M, Hirakawa M. KEGG for representation and analysis of molecular networks involving diseases and drugs. Nucleic Acids Res. 2010;38:D355–D360. doi: 10.1093/nar/gkp896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Caspi R, Altman T, Dale JM, Dreher K, Fulcher CA, Gilham F, Kaipa P, Karthikeyan AS, Kothari A, Krummenacker M, et al. The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases. Nucleic Acids Res. 2010;38:D473–D479. doi: 10.1093/nar/gkp875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jennen DG, Gaj S, Giesbertz PJ, van Delft JH, Evelo CT, Kleinjans JC. Biotransformation pathway maps in WikiPathways enable direct visualization of drug metabolism related expression changes. Drug Discov. Today. 2010;15:851–858. doi: 10.1016/j.drudis.2010.08.002. [DOI] [PubMed] [Google Scholar]
- 7.Eichelbaum M, Altman RB, Ratain M, Klein TE. New feature: pathways and important genes from PharmGKB. Pharmacogenet. Genomics. 2009;19:403. doi: 10.1097/FPC.0b013e32832b16ba. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Frolkis A, Knox C, Lim E, Jewison T, Law V, Hau DD, Liu P, Gautam B, Ly S, Guo AC, et al. SMPDB: the Small Molecule Pathway Database. Nucleic Acids Res. 2010;38:D480–D487. doi: 10.1093/nar/gkp1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schaefer CF, Anthony K, Krupa S, Buchoff J, Day M, Hannay T, Buetow KH. PID: the Pathway Interaction Database. Nucleic Acids Res. 2009;37:D674–D679. doi: 10.1093/nar/gkn653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Oda K, Kitano H. A comprehensive map of the toll-like receptor signaling network. Mol. Syst. Biol. 2006;2 doi: 10.1038/msb4100057. 2006.0015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Calzone L, Gelay A, Zinovyev A, Radvanyi F, Barillot E. A comprehensive modular map of molecular interactions in RB/E2F pathway. Mol. Syst. Biol. 2008;4:173. doi: 10.1038/msb.2008.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Matsuoka Y, Ghosh S, Kikuchi N, Kitano H. Payao: a community platform for SBML pathway model curation. Bioinformatics. 2010;26:1381–1383. doi: 10.1093/bioinformatics/btq143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cerami EG, Gross BE, Demir E, Rodchenkov I, Babur O, Anwar N, Schultz N, Bader GD, Sander C. Pathway Commons, a web resource for biological pathway data. Nucleic Acids Res. 2011;39:D685–D690. doi: 10.1093/nar/gkq1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kamburov A, Pentchev K, Galicka H, Wierling C, Lehrach H, Herwig R. ConsensusPathDB: toward a more complete picture of cell biology. Nucleic Acids Res. 2011;39:D712–D717. doi: 10.1093/nar/gkq1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Demir E, Cary MP, Paley S, Fukuda K, Lemer C, Vastrik I, Wu G, D'Eustachio P, Schaefer C, Luciano J, et al. The BioPAX community standard for pathway data sharing. Nat. Biotechnol. 2010;28:935–942. doi: 10.1038/nbt.1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Funahashi A, Matsuoka Y, Jouraku A, Morohashi M, Kikuchi N, Kitano H. CellDesigner 3.5: a versatile modeling tool for biochemical networks. Proc. IEEE. 2008;96:1254–1265. [Google Scholar]
- 17.Groenendyk J, Sreenivasaiah PK, Kim DH, Agellon LB, Michalak M. Biology of endoplasmic reticulum stress in the heart. Circ. Res. 2010;107:1185–1197. doi: 10.1161/CIRCRESAHA.110.227033. [DOI] [PubMed] [Google Scholar]
- 18.Davies MN, Meaburn EL, Schalkwyk LC. Gene set enrichment; a problem of pathways. Brief. Funct. Genomic. 2010;9:385–390. doi: 10.1093/bfgp/elq021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Le Novere N, Hucka M, Mi H, Moodie S, Schreiber F, Sorokin A, Demir E, Wegner K, Aladjem MI, Wimalaratne SM, et al. The systems biology graphical notation. Nat. Biotechnol. 2009;27:735–741. doi: 10.1038/nbt.1558. [DOI] [PubMed] [Google Scholar]
- 20.Lachmann A, Ma'ayan A. Lists2Networks: integrated analysis of gene/protein lists. BMC Bioinformatics. 2010;11:87. doi: 10.1186/1471-2105-11-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim SY, Volsky DJ. PAGE: parametric analysis of gene set enrichment. BMC Bioinformatics. 2005;6:144. doi: 10.1186/1471-2105-6-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goel R, Muthusamy B, Pandey A, Prasad TS. Human protein reference database and human proteinpedia as discovery resources for molecular biotechnology. Mol. Biotechnol. 2011;48:87–95. doi: 10.1007/s12033-010-9336-8. [DOI] [PubMed] [Google Scholar]
- 23.Ceol A, Chatr Aryamontri A, Licata L, Peluso D, Briganti L, Perfetto L, Castagnoli L, Cesareni G. MINT, the molecular interaction database: 2009 update. Nucleic Acids Res. 2010;38:D532–D539. doi: 10.1093/nar/gkp983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Massa MS, Chiogna M, Romualdi C. Gene set analysis exploiting the topology of a pathway. BMC Syst. Biol. 2010;4:121. doi: 10.1186/1752-0509-4-121. [DOI] [PMC free article] [PubMed] [Google Scholar]