Abstract
Lung cancer is the most common cause of cancer-related mortality with more than 1.4 million deaths per year worldwide. To search for significant somatic alterations in lung cancer, we analyzed, integrated and manually curated various data sets and literatures to present an integrated genomic database of non-small cell lung cancer (IGDB.NSCLC, http://igdb.nsclc.ibms.sinica.edu.tw). We collected data sets derived from hundreds of human NSCLC (lung adenocarcinomas and/or squamous cell carcinomas) to illustrate genomic alterations [chromosomal regions with copy number alterations (CNAs), gain/loss and loss of heterozygosity], aberrant expressed genes and microRNAs, somatic mutations and experimental evidence and clinical information of alterations retrieved from literatures. IGDB.NSCLC provides user friendly interfaces and searching functions to display multiple layers of evidence especially emphasizing on concordant alterations of CNAs with co-localized altered gene expression, aberrant microRNAs expression, somatic mutations or genes with associated clinicopathological features. These significant concordant alterations in NSCLC are graphically or tabularly presented to facilitate and prioritize as the putative cancer targets for pathological and mechanistic studies of lung tumorigenesis and for developing new strategies in clinical interventions.
INTRODUCTION
Cancer is the leading killer of human beings with more than 7.4 million deaths worldwide per year. Among them, lung cancer is the most common cause of cancer-related mortality in both men and women with over 1.4 million deaths annually (1). Lung cancer can be divided into two categories: small cell lung cancer (SCLC) and non-small cell lung cancer (NSCLC). Approximately 85% of total lung cancers are NSCLC that can be further classified into two major subtypes: squamous cell carcinoma (SCC) and adenocarcinoma (AD). Recent remarkable advances in identification of genetic alterations including p53, KRAS, EGFR, HER2, c-MET, LKB1, PIK3CA, BRAF and EML4-ALK not only provided mechanistic understanding of tumor growth and metastatic advantages but also served as targets for diagnosis and therapy (2–12). Moreover, a recent report suggested that high risk lung cancer individuals aged 55–74 years with heavy smoking history is associated with a 20% reduction in lung cancer mortality when computed tomography (CT) lung cancer screening is conducted in comparison with chest X-ray screening (http://www.cancer.gov/images/dsmb-nlst.pdf). Even with these recent advances, early screening of lung cancer is still inconsistent and the overall 5-year survival rate of lung cancer remains around 15% (13–15). Therefore, discovery of altered genes for early detection and as therapeutic targets is urgently needed to prolong the life of lung cancer patients.
Similar to many other cancers, lung cancer is a complex and heterogeneous genetic disease resulting from the accumulation of genetic and epigenetic alterations during multiple steps of tumor progression and metastasis (16,17). Depending on tumor subtype, ethnicity, gender and exposure of carcinogens (e.g. smoking, radon gas, asbestos and cooking oil fumes), accumulated alterations exploit different tumorigenic mechanisms resulting in aberrant activation of oncogenic signaling pathways and uncontrolled tumor growth and metastasis (18–20). To identify common cancer-associated alterations, application of high-throughput genomic technologies including DNA sequencing and high density microarrays allowed investigators to generate comprehensive genome-wide data sets of somatic mutations, copy number alterations (CNAs), transcriptomics and altered microRNA (miRNAs) expression of lung cancer genomes (21–33). In combination with clinical features, investigators were able to independently identified profiles from these genomic alterations for categorization of lung cancer subtypes, identification of cancer genes for tumorigenic studies, diagnostic and prognostic prediction of patient outcomes and development of various strategies to improve patient care.
To facilitate identification of common alterations that might embrace oncogenic or tumor suppressive genes in NSCLC, we curated numerous genomic data sets and established the integrated database IGDB.NSCLC. We graphically displayed multiple dimensional data of lung cancer somatic alterations including somatic mutations, CNAs, aberrant expression of genes and miRNAs, chromosomal gain and loss regions, frequent loss of heterozygosity (LOH) regions and experimental supports of alterations retrieved from lung cancer literatures. Altered gene expression in association with clinicopathological features and patient outcomes is also included for developing biomarkers in clinical managements. Although lung cancer is the most common cause of cancer-related mortality with rapid expansion of cancer genomic studies, limited efforts were made to integrate these genomic resources for a coherent and user-friendly presentation. A database HLungDB was reported as a helpful resource of human lung cancer research with integrated and networking analysis of lung cancer-related genes, proteins and miRNAs extracted manually from literatures (34). However, IGDB.NSCLC provides not only lung cancer genes and miRNAs from published reports but also analyzed various lung cancer genomic data sets for simultaneous illustrations of somatic alterations at genome, RNA, protein, function and application levels. The simultaneous illustrations provide multiple layers of evidence for investigators to detect and prioritize the common altered genes and miRNAs accessible to the scientific and medical communities. Moreover, we specifically emphasized on concordant alterations in altered NSCLC cancer genomes such as (i) up-regulated genes and miRNAs encoded in regions of genomic amplification; (ii) down-regulated genes and miRNAs encoded in regions of genomic deletion; (iii) mutation genes with concordant genomic alterations; and (iv) genes with significant association with clinical information encoded in regions of genomic alterations. These altered genes defined as significant genes and miRNAs in NSCLC were graphically and tabularly displayed in IGDB.NSCLC and should be prioritized to develop as useful targets for improvement of patient management.
DATABASE CONSTRUCTION
For integration of various genomic resources of NSCLC, we collected genomic data generated from primary tissue samples of lung AD and SCC separately (Supplementary Table S1). For altered gene expression performed in microarrays, we collected 15 data sets from 1099 AD, 295 SCC and 189 normal tissue samples from five different microarray platforms. An expression profiling data set (GSE14814) of 90 NSCLC samples with and without cisplatin/vinorelbine treatment was included for revealing potential prognostic markers in NSCLC (35). We also included a reported pair data set of 193 lung AD samples with genomic alteration performed in 44 K Agilent CGH array and expression alteration in Affymetrix U133A and U133 2.0 arrays for concordant alterations from the same samples (36).
For processing Affymetrix gene expression data sets, the raw data (CEL files) were normalized by MAS 5.0 (an affy package from Bioconductor/R at http://www.bioconductor.org/packages/release/bioc/html/affy.html) (37). Quality control (QC) results were performed by the simpleaffy and affyQCReport packages (http://www.bioconductor.org/packages/release/bioc/html/affyQCReport.html and http://www.bioconductor.org/packages/release/bioc/html/simpleaffy.html) with three stringent QC criteria for inclusion of a microarray data: (i) the scaling factor of a given sample should be within two standard deviations of mean of the same array platform, (ii) the present calls of a given sample should be >25% and (iii) the 3′/5′ GAPDH ratios of a given sample should be <3 (38). After these QC process, 46 samples were eliminated (HG_U95A: 22 samples, HG-U133A: 2 samples and HG-U133_Plus_2: 22 samples). The data sets from the same platform were normalized using the normalize.quantiles in R. Finally, all probeset intensity value was transformed to log 2 value. For data performed in two color microarrays (Agilent and Stanford data sets), expression profiles were downloaded form GEO website directly. The log 10 ratio of the Agilent data set was transformed to log 2 ratio. The differential expression genes for each platform were ranked on moderated t-statistics and selected with P < 0.01 under the Bayesian adjusted t-statistics from the linear models for microarray data (limma) package (39).
For detection of CNAs, we downloaded three data sets of 191 AD, 117 SCC and 271 normal tissue samples performed in Affymetrix GeneChip single nucleotide polymorphism (SNP) arrays and analyzed by dChip software (40). In brief, CEL format data are normalized using invariant set normalization algorithms and then normalized-within-chip intensity data are generated based on the reference data set of 50 normal individuals genotyped in the same platform. Based on these signal values, the raw copy number for an SNP in a sample is computed as: [log 2(intensity of SNP/mean of intensity of reference × 2)]. A window size of three SNPs is then applied for median smoothing method and to infer raw copy number (ICN) (41). We defined the amplified regions with ICN more than three and deleted regions with ICN less than one (42). For altered expression of miRNAs, two data sets of 193 AD and 137 SCC tissue samples were collected and the miRNAs expression was analyzed by calculating the mean (±standard deviation) of the log 2 ratios of tumor/non-tumor adjacent tissues. For analysis of arrayed comparative genome hybridization (aCGH) data, we obtained two data sets of 70 SCC tissue samples and analyzed them using the cghMCR package (http://www.bioconductor.org/packages/2.4/bioc/html/cghMCR.html). The gain and loss regions were calculated based on the SGOL (Segment Gain Or Loss) scores by calculating the summations of all the positive values over a threshold of copy number (CN) >3 and all the negative values below a threshold of CN < 1.2, respectively. In addition, we also integrated at least 1112 somatic mutation genes of NSCLC from COSMIC and literatures, 214 lung cancer genes with experimental evidence, 131 genes with association with clinicopathological features of NSCLC and other genomic alterations such as LOH and minimum region of alterations from literatures and our unpublished data.
RESULTS
Multiple levels of somatic alterations in NSCLC
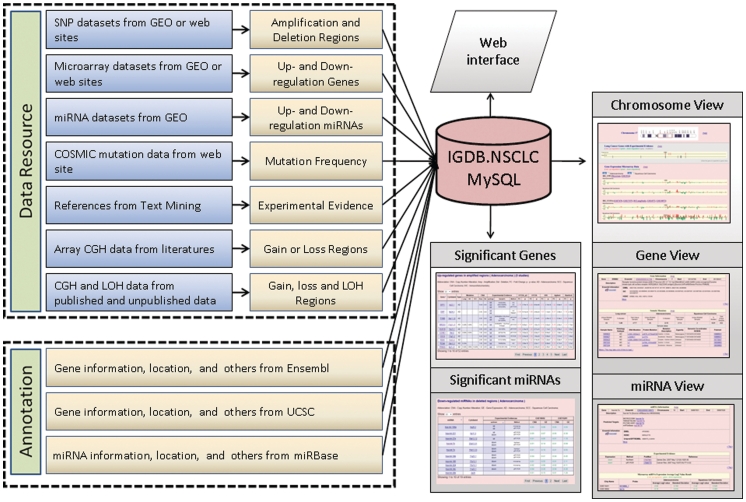
To facilitate identification of cancer-associated somatic altered genes, we aimed to provide lines of evidence for investigators to prioritize these genes for mechanistic studies and clinical applications in NSCLC. The integrated framework of IGDB.NSCLC is constructed based on the physical map of human genome sequence from Ensembl with various integrated somatic alterations alongside (Figure 1). Users could apply various searching terms (gene, marker, cytogentic location and other key words) for displaying the altered data in lung AD, SCC and/or combined NSCLC. Three major illustrations including chromosome view, gene view and miRNA view are provided for simultaneous displays of concordant alterations in the same interface. In the chromosome view, the interface demonstrated the comprehensive and integrated alterations in relation to NSCLC genes and regions including evidence of experimental support, somatic mutation, altered gene expression, CNAs, LOH and other chromosomal alterations. We provided ERBB2, EGFR and MET as tutorial examples (please see our quick examples of chromosome view). In gene view, we presented the details of aberrant information of the selected gene including (i) mutation frequency and details of mutation changes; (ii) frequency of altered gene expression supported by experimental evidence such as immunohistochemistry (IHC), RT–PCR or western blot analysis and in association with clinical information; (iii) the fold changes and the statistic significance of altered expression of each probe set in a selected gene from a given microarray platform; (iv) the distribution plots of the altered expressing gene in a set of multiple NSCLC tissues shown in log 2 ratios of tumor/non-tumor adjacent tissues; and (v) the publication status of the aberrant gene in various cancers provided in the cancer gene index and the literature (see our quick examples of gene view). In miRNA view, we also provided details of the aberrant expression of a miRNA as similar to that of gene view in addition to external links for predicting miRNA target genes in MICROCOSM (43), TARGETSCAN (44), PICTAR-VERT (45) and miRTarBase (46). We provided three miRNAs including hsa-let-7e, hsa-mir-17 and hsa-mir-31 as tutorial examples (see our quick examples of miRNA view).
Figure 1.
The framework of IGDB.NSCLC.
Concordant somatic alterations in NSCLC
To further reveal significant cancer-associated genes and miRNAs involved in tumor progression of NSCLC, we applied CNA (amplification: Inferred copy number ICN >3 and deletion: ICN <1) as the framework and searched for concordant existence of (i) genes with altered expression, (ii) miRNA with altered expression, (iii) genes with common somatic mutations and (iv) genes associated with clinicopathological features from literatures. For genes with altered expression existing in regions of CNA, we found 27 up-regulated genes located in amplified regions and seven down-regulated genes residing in deleted regions in AD; and 23 up-regulated genes in amplified regions and 13 down-regulated genes in deleted regions in SCC (Figure 2 and Supplementary Table S2). Some of these genes were previously identified as altered expression genes in NSCLC for validation but majority of these genes remain unknown in the tumorigenesis of NSCLC. For miRNAs with altered expression residing in CNA regions, we identified 21 up-regulated miRNAs located in the amplified regions and 19 down-regulated miRNAs in the deleted regions in AD, and 19 up-regulated miRNAs in the amplified regions and 20 down-regulated miRNAs in the deleted regions in SCC (Supplementary Figure S1 and Supplementary Table S3). Similarly, we found that some of these aberrant miRNAs were validated by previous reports but majority of them are novel significant miRNAs in the tumor formation of NSCLC.
Figure 2.
The graphic integration of CNAs with altered expression genes in lung AD and SCC. The red lines represent the amplification regions for CNA and up-regulation genes. The green lines stand for the deletion regions for CNA and down-regulated genes. The lines are relatively corresponding to the physical position along the chromosomes.
We also examined the concordance of genes with common somatic mutations (at least mutations in five cases) that resides in the CNA regions of NSCLC. A total of 1112 genes with somatic mutations including 386 in AD and 55 in SCC were downloaded from COSMIC database (47). Majority of these somatic mutations occurred only once in NSCLC tissues except 43 genes in AD and seven genes in SCC conferred at least five independent somatic mutations. Interestingly, majority of these common mutated genes including 33/43 (76.7%) in AD and 6/7 (85.7%) in SCC are co-localized with CNA regions in NSCLC (Table 1). Our results further demonstrated that genes with frequent somatic mutations are commonly affiliated with chromosomal alterations involved in the tumorigenesis of NSCLC.
Table 1.
Common mutated genes in copy number alteration regions of NSCLC
| Non-small cell lung carcinoma | Alteration | Genes with at least five somatic mutations |
|---|---|---|
| AD | Mutated genes in amplified regions (30 genes) | APC, BRAF, CDC42BPA, CDKN2A, CTNNB1, EGFR, EPHA3, EPHA5, EPHA7, EPHB6, FGFR4, FLT1, INSRR, JAK2, KDR, KIAA1804, KRAS, LMTK2, MET, NRAS, NTRK1, PAK7, PDGFRA, PIK3C3, PIK3CA, PIK3CG, PRKDC, RB1, ROBO2, TERT |
| Mutated genes in deleted regions | NOTCH1, PTEN, TP53 | |
| SCC | Mutated genes in amplified regions | BRAF, CDKN2A, EGFR, KRAS, PIK3CA |
| Mutated genes in deleted regions | TP53 |
To further examine the link between clinicopathological features with somatic alterations, we collected 309 papers with 125 genes in AD and 105 genes in SCC validated by experimental evidence such as IHC, RT–PCR and western analysis for confirmation of altered expression in NSCLC. Among them, 62 (62/125, 49.6%) genes in AD and 54 (54/105, 51.4%) genes in SCC extracted from 159 papers were shown to associate with 22 clinicopathological features and to be located in the CNA regions (Supplementary Table S4). Our results suggested that NSCLC genes associated with clinicopathological features commonly (∼50%) reside in the CNA regions, reflecting the genetic effects participated in pathological changes of tumor formation in NSCLC.
DISCUSSION
As far as we know, IGDB.NSCLC is the largest integration of lung cancer genomic resources providing multiple levels of evidence to search for the concordantly altered targets and to prioritize putative NSCLC genes for future studies. In addition to the database with various searching options and user-friendly interfaces, we provided concordant somatic alterations based on the genome-wide CNAs data with co-localization of altered gene expression, aberrant miRNA expression, somatic mutation and genes in association with clinicopathological features. The high concordance of CNA data with these somatic alterations and clinical features in IGDB.NSCLC suggested the quality of data integration with experimental validations, the heterogeneity of genetic pathways to NSCLC and the important roles of genomic alterations involved in tumor formation of NSCLC. The future development of integrating other NSCLC resources into IGDB.NSCLC will focus on increasing the data of clinicopathological features for dissecting the altered genes involved in tumor progression and on somatic alteration data from next-generation sequencing results. In conclusion, IGDB.NSCLC is an invaluable resource for selecting putative cancer genes in NSCLC to better understand the heterogeneous tumorigenic mechanisms and for developing useful strategies in clinical applications to prolong the life of lung cancer patients.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–4, Supplementary Figure 1.
FUNDING
National Research Program for Genomic Medicine (NSC98-3112-B-001-004, NSC98-3112-B-001-031); National Research Program for Biopharmaceuticals, National Science Council, Taiwan (NSC100-2325-B-001-012). Funding for open access charge: Institute of Biomedical Sciences, Academia Sinica, Taiwan.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The following authors contributed toward this study: database construction: S.K. and C.K.S.; data analysis and interpretation: S.K., D.L.G., C.M.H. and C.F.C.; drafting of the manuscript and study supervision: W.H.S., C.F.C., C.H.L. and Y.S.J.
REFERENCES
- 1. WHO. World Health Organization. “Cancer”. http://www.who.int/mediacentre/factsheets/fs297/en/. (3 March 2010, date last accessed)
- 2.Brose MS, Volpe P, Feldman M, Kumar M, Rishi I, Gerrero R, Einhorn E, Herlyn M, Minna J, Nicholson A, et al. BRAF and RAS mutations in human lung cancer and melanoma. Cancer Res. 2002;62:6997–7000. [PubMed] [Google Scholar]
- 3.Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, Brannigan BW, Harris PL, Haserlat SM, Supko JG, Haluska FG, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N. Engl. J. Med. 2004;350:2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 4.Ma PC, Jagadeeswaran R, Jagadeesh S, Tretiakova MS, Nallasura V, Fox EA, Hansen M, Schaefer E, Naoki K, Lader A, et al. Functional expression and mutations of c-Met and its therapeutic inhibition with SU11274 and small interfering RNA in non-small cell lung cancer. Cancer Res. 2005;65:1479–1488. doi: 10.1158/0008-5472.CAN-04-2650. [DOI] [PubMed] [Google Scholar]
- 5.Naoki K, Chen TH, Richards WG, Sugarbaker DJ, Meyerson M. Missense mutations of the BRAF gene in human lung adenocarcinoma. Cancer Res. 2002;62:7001–7003. [PubMed] [Google Scholar]
- 6.Paez JG, Janne PA, Lee JC, Tracy S, Greulich H, Gabriel S, Herman P, Kaye FJ, Lindeman N, Boggon TJ, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 7.Samuels Y, Velculescu VE. Oncogenic mutations of PIK3CA in human cancers. Cell Cycle. 2004;3:1221–1224. doi: 10.4161/cc.3.10.1164. [DOI] [PubMed] [Google Scholar]
- 8.Samuels Y, Wang Z, Bardelli A, Silliman N, Ptak J, Szabo S, Yan H, Gazdar A, Powell SM, Riggins GJ, et al. High frequency of mutations of the PIK3CA gene in human cancers. Science. 2004;304:554. doi: 10.1126/science.1096502. [DOI] [PubMed] [Google Scholar]
- 9.Sanchez-Cespedes M, Parrella P, Esteller M, Nomoto S, Trink B, Engles JM, Westra WH, Herman JG, Sidransky D. Inactivation of LKB1/STK11 is a common event in adenocarcinomas of the lung. Cancer Res. 2002;62:3659–3662. [PubMed] [Google Scholar]
- 10.Santos E, Martin-Zanca D, Reddy EP, Pierotti MA, Della Porta G, Barbacid M. Malignant activation of a K-ras oncogene in lung carcinoma but not in normal tissue of the same patient. Science. 1984;223:661–664. doi: 10.1126/science.6695174. [DOI] [PubMed] [Google Scholar]
- 11.Stephens P, Hunter C, Bignell G, Edkins S, Davies H, Teague J, Stevens C, O'Meara S, Smith R, Parker A, et al. Lung cancer: intragenic ERBB2 kinase mutations in tumours. Nature. 2004;431:525–526. doi: 10.1038/431525b. [DOI] [PubMed] [Google Scholar]
- 12.Takahashi T, Nau MM, Chiba I, Birrer MJ, Rosenberg RK, Vinocour M, Levitt M, Pass H, Gazdar AF, Minna JD. p53: a frequent target for genetic abnormalities in lung cancer. Science. 1989;246:491–494. doi: 10.1126/science.2554494. [DOI] [PubMed] [Google Scholar]
- 13.Bach PB. Inconsistencies in findings from the early lung cancer action project studies of lung cancer screening. J. Natl. Cancer Inst. 2011;103:1002–1006. doi: 10.1093/jnci/djr202. [DOI] [PubMed] [Google Scholar]
- 14.Van't Westeinde SC, van Klaveren RJ. Screening and early detection of lung cancer. Cancer J. 2011;17:3–10. doi: 10.1097/PPO.0b013e3182099319. [DOI] [PubMed] [Google Scholar]
- 15.Jemal A, Siegel R, Ward E, Hao Y, Xu J, Thun MJ. Cancer statistics, 2009. CA Cancer J. Clin. 2009;59:225–249. doi: 10.3322/caac.20006. [DOI] [PubMed] [Google Scholar]
- 16.Agullo-Ortuno MT, Lopez-Rios F, Paz-Ares L. Lung cancer genomic signatures. J. Thorac. Oncol. 2010;5:1673–1691. doi: 10.1097/JTO.0b013e3181f1900e. [DOI] [PubMed] [Google Scholar]
- 17.Risch A, Plass C. Lung cancer epigenetics and genetics. Int. J. Cancer. 2008;123:1–7. doi: 10.1002/ijc.23605. [DOI] [PubMed] [Google Scholar]
- 18.Chaffer CL, Weinberg RA. A perspective on cancer cell metastasis. Science. 2011;331:1559–1564. doi: 10.1126/science.1203543. [DOI] [PubMed] [Google Scholar]
- 19.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 20.Sun S, Schiller JH, Gazdar AF. Lung cancer in never smokers—a different disease. Nat. Rev. Cancer. 2007;7:778–790. doi: 10.1038/nrc2190. [DOI] [PubMed] [Google Scholar]
- 21.Puissegur MP, Mazure NM, Bertero T, Pradelli L, Grosso S, Robbe-Sermesant K, Maurin T, Lebrigand K, Cardinaud B, Hofman V, et al. miR-210 is overexpressed in late stages of lung cancer and mediates mitochondrial alterations associated with modulation of HIF-1 activity. Cell Death Differ. 2011;18:465–478. doi: 10.1038/cdd.2010.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Landi MT, Zhao Y, Rotunno M, Koshiol J, Liu H, Bergen AW, Rubagotti M, Goldstein AM, Linnoila I, Marincola FM, et al. MicroRNA expression differentiates histology and predicts survival of lung cancer. Clin. Cancer Res. 2010;16:430–441. doi: 10.1158/1078-0432.CCR-09-1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bass AJ, Watanabe H, Mermel CH, Yu S, Perner S, Verhaak RG, Kim SY, Wardwell L, Tamayo P, Gat-Viks I, et al. SOX2 is an amplified lineage-survival oncogene in lung and esophageal squamous cell carcinomas. Nat. Genet. 2009;41:1238–1242. doi: 10.1038/ng.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boelens MC, Kok K, van der Vlies P, van der Vries G, Sietsma H, Timens W, Postma DS, Groen HJ, van den Berg A. Genomic aberrations in squamous cell lung carcinoma related to lymph node or distant metastasis. Lung Cancer. 2009;66:372–378. doi: 10.1016/j.lungcan.2009.02.017. [DOI] [PubMed] [Google Scholar]
- 25.Broet P, Camilleri-Broet S, Zhang S, Alifano M, Bangarusamy D, Battistella M, Wu Y, Tuefferd M, Regnard JF, Lim E, et al. Prediction of clinical outcome in multiple lung cancer cohorts by integrative genomics: implications for chemotherapy selection. Cancer Res. 2009;69:1055–1062. doi: 10.1158/0008-5472.CAN-08-1116. [DOI] [PubMed] [Google Scholar]
- 26.Ding L, Getz G, Wheeler DA, Mardis ER, McLellan MD, Cibulskis K, Sougnez C, Greulich H, Muzny DM, Morgan MB, et al. Somatic mutations affect key pathways in lung adenocarcinoma. Nature. 2008;455:1069–1075. doi: 10.1038/nature07423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shedden K, Taylor JM, Enkemann SA, Tsao MS, Yeatman TJ, Gerald WL, Eschrich S, Jurisica I, Giordano TJ, Misek DE, et al. Gene expression-based survival prediction in lung adenocarcinoma: a multi-site, blinded validation study. Nat. Med. 2008;14:822–827. doi: 10.1038/nm.1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weir BA, Woo MS, Getz G, Perner S, Ding L, Beroukhim R, Lin WM, Province MA, Kraja A, Johnson LA, et al. Characterizing the cancer genome in lung adenocarcinoma. Nature. 2007;450:893–898. doi: 10.1038/nature06358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Raponi M, Zhang Y, Yu J, Chen G, Lee G, Taylor JM, Macdonald J, Thomas D, Moskaluk C, Wang Y, et al. Gene expression signatures for predicting prognosis of squamous cell and adenocarcinomas of the lung. Cancer Res. 2006;66:7466–7472. doi: 10.1158/0008-5472.CAN-06-1191. [DOI] [PubMed] [Google Scholar]
- 30.Takeuchi T, Tomida S, Yatabe Y, Kosaka T, Osada H, Yanagisawa K, Mitsudomi T, Takahashi T. Expression profile-defined classification of lung adenocarcinoma shows close relationship with underlying major genetic changes and clinicopathologic behaviors. J. Clin. Oncol. 2006;24:1679–1688. doi: 10.1200/JCO.2005.03.8224. [DOI] [PubMed] [Google Scholar]
- 31.Tseng RC, Chang JW, Hsien FJ, Chang YH, Hsiao CF, Chen JT, Chen CY, Jou YS, Wang YC. Genomewide loss of heterozygosity and its clinical associations in non small cell lung cancer. Int. J. Cancer. 2005;117:241–247. doi: 10.1002/ijc.21178. [DOI] [PubMed] [Google Scholar]
- 32.Garber ME, Troyanskaya OG, Schluens K, Petersen S, Thaesler Z, Pacyna-Gengelbach M, van de Rijn M, Rosen GD, Perou CM, Whyte RI, et al. Diversity of gene expression in adenocarcinoma of the lung. Proc. Natl. Acad. Sci. USA. 2001;98:13784–13789. doi: 10.1073/pnas.241500798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bhattacharjee A, Richards WG, Staunton J, Li C, Monti S, Vasa P, Ladd C, Beheshti J, Bueno R, Gillette M, et al. Classification of human lung carcinomas by mRNA expression profiling reveals distinct adenocarcinoma subclasses. Proc. Natl. Acad. Sci. USA. 2001;98:13790–13795. doi: 10.1073/pnas.191502998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang L, Xiong Y, Sun Y, Fang Z, Li L, Ji H, Shi T. HLungDB: an integrated database of human lung cancer research. Nucleic Acids Res. 2010;38:D665–D669. doi: 10.1093/nar/gkp945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhu CQ, Ding K, Strumpf D, Weir BA, Meyerson M, Pennell N, Thomas RK, Naoki K, Ladd-Acosta C, Liu N, et al. Prognostic and predictive gene signature for adjuvant chemotherapy in resected non-small-cell lung cancer. J. Clin. Oncol. 2010;28:4417–4424. doi: 10.1200/JCO.2009.26.4325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chitale D, Gong Y, Taylor BS, Broderick S, Brennan C, Somwar R, Golas B, Wang L, Motoi N, Szoke J, et al. An integrated genomic analysis of lung cancer reveals loss of DUSP4 in EGFR-mutant tumors. Oncogene. 2009;28:2773–2783. doi: 10.1038/onc.2009.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gautier L, Cope L, Bolstad BM, Irizarry RA. affy—analysis of Affymetrix GeneChip data at the probe level. Bioinformatics. 2004;20:307–315. doi: 10.1093/bioinformatics/btg405. [DOI] [PubMed] [Google Scholar]
- 38.Tumor Analysis Best Practices Working Group Expression profiling—best practices for data generation and interpretation in clinical trials. Nat. Rev. Genet. 2004;5:229–237. doi: 10.1038/nrg1297. [DOI] [PubMed] [Google Scholar]
- 39.Wettenhall JM, Smyth GK. limmaGUI: a graphical user interface for linear modeling of microarray data. Bioinformatics. 2004;20:3705–3706. doi: 10.1093/bioinformatics/bth449. [DOI] [PubMed] [Google Scholar]
- 40.Li C, Wong WH. Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc. Natl. Acad. Sci. USA. 2001;98:31–36. doi: 10.1073/pnas.011404098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shiau CK, Gu DL, Chen CF, Lin CH, Jou YS. IGRhCellID: integrated genomic resources of human cell lines for identification. Nucleic Acids Res. 2011;39:D520–D524. doi: 10.1093/nar/gkq1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen CF, Hsu EC, Lin KT, Tu PH, Chang HW, Lin CH, Chen YJ, Gu DL, Wu JY, Chen YT, et al. Overlapping high-resolution copy number alterations in cancer genomes identified putative cancer genes in hepatocellular carcinoma. Hepatology. 2010;52:1690–1701. doi: 10.1002/hep.23847. [DOI] [PubMed] [Google Scholar]
- 43.Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. miRBase: tools for microRNA genomics. Nucleic Acids Res. 2008;36:D154–D158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 45.Krek A, Grun D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, Stoffel M, et al. Combinatorial microRNA target predictions. Nat. Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 46.Sethupathy P, Corda B, Hatzigeorgiou AG. TarBase: A comprehensive database of experimentally supported animal microRNA targets. RNA. 2006;12:192–197. doi: 10.1261/rna.2239606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Forbes SA, Tang G, Bindal N, Bamford S, Dawson E, Cole C, Kok CY, Jia M, Ewing R, Menzies A, et al. COSMIC (the Catalogue of Somatic Mutations in Cancer): a resource to investigate acquired mutations in human cancer. Nucleic Acids Res. 2010;38:D652–D657. doi: 10.1093/nar/gkp995. [DOI] [PMC free article] [PubMed] [Google Scholar]